
A novel missense creatine mutant of CaBP4, c.464G>A (p.G155D), associated with autosomal dominant nocturnal frontal lobe epilepsy (ADNFLE), reduces the expression of CaBP4
Introduction
Autosomal dominant nocturnal frontal lobe epilepsy (ADNFLE) is a type of epilepsy syndrome characterized by clusters of nocturnal sleep-related motor seizures with onset at any age and inherited in an autosomal dominant manner (1,2). ADNFLE is the first disease in which a monogenic-inherited focal epilepsy syndrome has a clearly related pathogenic gene (3). At present, the known pathogenic genes are mainly neuronal acetylcholine receptor (nAChR)-related genes (1,3-7). In recent years, many genes, such as KCNT1, DEPDC5, and CRH, have been confirmed to be associated with ADNFLE (8-10), but the genotypes for a significant proportion of patients with this disease are still unknown (1). In the previous study of our group, whole-gene exome sequencing was used for analysis of blood samples collected from ADNFLE patients and family members. And through this, our research group previously identified a novel missense mutation, c.464G>A (p.G155D), in Ca2+-binding protein 4 (CaBP4) in a Chinese pedigree with ADNFLE and proved that mutations in the CaBP4 gene might be related to ADNFLE (2).
CaBP4 is a member of the Ca2+-binding protein (CaBP) family characterized by four EF-hand motifs encoded by the CaBP4 gene. CaBP is a subfamily of the calmodulin (CaM)-like neuronal CaBPs that modulate voltage-dependent Ca2+ channels (VDCCs) and inositol triphosphate (11). The mutations of CaBP4, which are reported primarily in congenital stationary night blindness (CSNB), probably cause the disease by influencing the function of CaBP4, which modulates the activation of VDCCs in the photoreceptor synaptic terminals (11).
Recent studies suggest that CaBP4 can regulate the activity of the Cav1.4 L-type Ca2+ channels. Park et al. showed that CaBP4 is part of the Cav1.4 channel complex in the retina and is implicated in the modulation of voltage-dependent Cav1.4 activation (12). Shaltiel et al. provided evidence that CaBP4 affects Cav1.4 by structural interference with the binding of the inhibitor of Ca2+-dependent inactivation (ICDI) domain to the C terminus of Cav1.4 (13). Cav1.4 is a member of the voltage-gated calcium channels (VGCCs). VGCCs are widely expressed in the mammalian central nervous system (CNS). Most variations of gene coding VGCCs have long been implicated as a susceptibility factor in the pathogenesis of idiopathic generalized epilepsy (14). If the c.464G>A (p.G155D) mutation in the CaBP4 gene influences the expression level of CaBP4 in the neurons, this could result in an imbalance in the depolarizing and excitatory effects of VGCCs expressed in the CNS.
In this study, we established human neuron models transfected with recombinant CaBP4 plasmid. By observing the survival rate of the human neurons, we found that the c.464G>A (p.G155D) mutation in the CaBP4 gene can influence the function of CaBP4. Moreover, analysis of the protein levels indicated that the c.464G>A (p.G155D) mutation in the CaBP4 gene reduced the protein expression of CaBP4. Our results confirm the mechanism of the missense mutation c.464G>A (p.G155D) in the CaBP4 gene in influencing CaBP4 expression and the role of CaBP4 mutations in the pathogenesis of ADNFLE.
The p.G155D mutation in CaBP4 mentioned in this paper was discovered in the ADNFLE family by our group on the first time. It provides a new idea for the study of the pathogenesis of ADNFLE.
We present the following article in accordance with the MDAR reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-22-54/rc).
Methods
In vitro mutagenesis
Full-length human CaBP4 cDNA construct and CaBP4 cDNA construct with the c.464G>A (p.G155D) mutation (subcloned in a pEGFP-N1 vector marked by green fluorescent protein) were purchased from Guangzhou IGE Biotechnology Ltd. (Guangzhou, China). The products were transformed in E. coli cells, and the plasmid DNA was isolated and purified using a HiPure Rapid Midiprep Plasmid Kit (Guangzhou, China). The purified plasmid DNA was sequenced using an ABI 3730XL sequencing platform (Shanghai, China).
Transient transfection of wild type, c.464G>A (p.G155D) CaBP4 and pEGFP-N1 in the human neuronal cell line
The human neuronal cell line was bought from Shanghai Baiyi Biotechnology Center (Shanghai, China). The human neuronal cells were seeded into 6-well plates and cultured for 48 hours. The CaBP4 plasmid DNA was mixed with the transfection reagent, EndofectinTM Max (GeneCopoeia, EF004). The reagent complex was then added to the human neuronal cells in a serum-free medium and incubated at 37 ℃ for 24 hours.
Collection of mRNA and protein from the cell lysate
The cells were washed with cold PBS (phosphate buffered saline) and lysed using a cell lysis buffer. The cell lysates were centrifuged at 12,000 rpm for 15 minutes at 4 ℃. The mRNA was collected from the supernatant, and its concentration was estimated using the Nano-100. The protein concentration was estimated using a BCA reagent (Shanghai, China).
Real-time polymerase chain reaction (RT-PCR)
The PCR samples had a final volume of 20 mL containing 1× SYBR Green Supermix (BioRad, Shanghai, China), 1 primer, and 5 mL cDNA (1:200). The primers used were as follows: H-CaBP4-F (5'-TTCGAGGAGTTTGACACTGACC-3'), H-CaBP4-R (5'-CCCATGCGCATCTTGATGTG-3'), H-GAPDH-F (5'-GAGTCAACGGATTTGGTCGT-3'), and H-GAPDH-R (5'-GACAAGCTTCCCGTTCTCAG-3'). Amplification was performed on a real-time quantitative PCR (ABI, StepOne Plus, America). For each run, a melting curve analysis was performed to confirm the specificity of the amplification. The results were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as the reference gene.
Western blot
Cell lysates (30 µg) of c.464G>A (p.G155D) as well as wild type and pEGFP-N1 were electrophoresed in a 10% concentration of polyacrylamide gel at 100 V for 2 hours. The proteins were electrophoretically transferred to a nitrocellulose membrane and blocked with 5% skimmed milk powder in PBS. The membrane was washed and incubated with polyclonal rabbit anti-green fluorescent protein antibody (1:1,000) at 4 ℃ overnight. The membrane was washed twice by TBST [TBS (Tris hydrochloride) +Tween] and incubated with biotinylated anti-rabbit IgG (1:3,000), marked by horseradish peroxidase (HRP) for 2 hours. The amount of protein for each band was then determined by chemiluminescence development.
Statistical analysis
The data in the paper were expressed in the form of mean ± standard deviation and were statistically analyzed with one-way analysis of variance (ANOVA) by SPSS26.0 software (IBM, America). The pictures were analyzed and drawn by GraphPad Prism 5.0 software (GraphPad Software, America). The raw data obtained in the RT-PCR experimental part were processed by the 2−ΔΔCt method and then statistically analyzed. The bands obtained in the experimental part were counted by immunofluorescence blotting and analyzed by ImageJ software to obtain the gray value of the bands, and then statistical analysis was performed.
Results
Plasmid gene sequencing
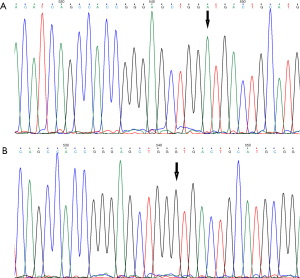
The extracted mutant recombinant plasmid (pEGFP-N1-CaBP4-p.G155D) and wild recombinant plasmid (pEGFP-N1-CaBP4) were verified by DNA sequencing on an ABI 3730XL sequencing platform, and the base sequence was found to be consistent with the designed sequence. The wild-type and mutant-type sequences of the mutant bases of the plasmid are shown in Figure 1A,1B. The plasmid was constructed successfully.
Transfection efficiency
We transfected the constructed empty plasmid (pEGFP-N1), wild recombinant plasmid (pEGFP-N1-CaBP4), and mutant recombinant plasmid (pEGFP-N1-CaBP4-p.G155D) into the human neuronal cell lines. They were divided into a blank control group, a wild control group, and a mutation experimental group. After 24 hours of culture, the green fluorescence expression of the human neuron cells in each group was observed by a light microscope and an inverted fluorescence microscope, and the transfection efficiency was calculated. The calculation results showed that the transfection efficiency of the blank control group was 20.42%±0.72%, the transfection efficiency of the wild control group was 15.24%±0.70% and the transfection efficiency of the mutation experiment group was 17.78%±0.72%.
Real-time PCR
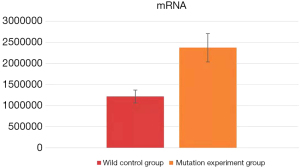
Real-time fluorescence quantitative detection was performed on the mRNA extracts of the human neuronal cells after transfection. The data obtained from the wild control group (human neuronal cells transfected with pEGFP-N1-CaBP4 wild recombinant plasmid) and the mutant experimental group (transfected with pEGFP)-N1-CaBP4-p.G155D mutant recombinant plasmid human neuron cells) were entered into Microsoft Excel for processing, using the 2−ΔΔCt method for data analysis. SPSS v.26.0 software (IBM, America) was used for a one-way analysis of variance (ANOVA), with a P value <0.05 assumed to indicate a statistically significant difference. The average value of 2−ΔΔCt in the wild control group was 1,216,779.27±152,976.26 compared with 2,372,403.48±335,942.63 in the mutation experiment group (P=0.011; Figure 2), indicating a statistically significant difference between the two groups. Specifically, the mRNA expression of the mutation experiment group was significantly higher than that of the wild control group. From this analysis, we concluded that the p.G155D mutation in the CaBP4 gene does not reduce CaBP4 mRNA expression and even has a tendency to increase its expression.
Western blot analysis
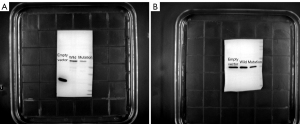
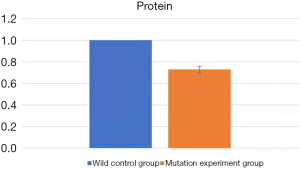
After transfection, the protein extracts of the human neuron cells were detected by western blot. ImageJ software was used to analyze the gray level of each band in the obtained gel photos (shown in Figure 3), and the housekeeping protein GAPDH was used as the internal reference protein. After the gray protein levels of the mutant experimental group (pEGFP-N1-CaBP4-p.G155D) and the wild control group (pEGFP-N1-CaBP4) were obtained, SPSS v.26.0 software (IBM, America) was used to apply a one-way ANOVA to analyze the standardized data with an assumed P value <0.05 indicating statistical significance. The average protein gray ratio of the control group was 1.000000 compared with 0.728050±0.087525 for the experimental group (P=0.01; Figure 4). The difference between the two groups was significant. Specifically, the protein expression of the experimental mutation group (pEGFP-N1-CaBP4-p.G155D) was significantly lower than that of the wild control group (pEGFP-N1-CaBP4). The CaBP4 gene p.G155D mutation may affect the stability of the protein, leading to the hydrolysis of the generated CaBP4 in the cell damage.
Discussion
ADNFLE is a type of epilepsy syndrome characterized by clusters of nocturnal sleep-related motor seizures. It is inherited in an autosomal dominant manner, with onset occurring at any stage of life (15,16). ADNFLE is the first disease for which a causative gene is clearly identified, with apparent genetic heterogeneity (3). Previous research has identified six pathogenic genes implicated in the disease (1,8,9,17). To date, the related pathogenic genes appear to be mainly concentrated in nAChR-related genes. GATOR, which corresponds to the protein subunit DEP domain containing 5, GATOR1 subcomplex subunit and DEPDC5-related genes, and the potassium channel subfamily T member 1 (potassium sodium-activated channel subfamily T member 1, KCNT1) genes are the currently known nAChR-related pathogenic genes. However, the genotype of approximately 70% of ADNFLE families remains unclear (18).
In the preliminary research work of this group, whole-genome exome sequencing technology and the Sanger sequencing method were applied to a family with ADNFLE. In this family of four generations, including 46 individuals and 11 affected patients, we found a new mutant genotype of the CaBP4 gene, p.G155D (2). We hypothesized that the mutation might be a new pathogenic gene in ADNFLE, and by using in vitro cell patch-clamp detection technology, we confirmed that the CaBP4 gene p.G155D mutation could cause action potentials on hippocampal neuron cell membranes. A significant increase in frequency leads to the onset of ADNFLE, but its mechanism of action has remained unclear (17).
In the process of reviewing and analyzing previous reports of CaBP4, we found that CaBP4 plays an important role in the regulation of cell signal transduction and the formation of synapses in neuronal cells. The CaBP4 gene is located at 11q13.2, is 866 bp in length, has six exons, encodes Calnexin 4, and has a total of 275 amino acids. As a CaBP, CaBP4 contains four EF-hand domains, grouped into two domains connected by a central linker. Contrary to CaM, CaBP4 contains 100 non-conserved amino acids upstream of the EF-hand domain in the N-terminal region. Another notable feature is that the second EF-hand domain lacks the critical residues required for high-affinity Ca2+-binding. According to current research, CaBP4 is mainly expressed in the rods and cones of the retina, the omentum, and the endothelial cells of the cochlea. It regulates the influx of Ca2+ to control the neurotransmitters at the synaptic terminals of photoreceptors and auditory receptors to control vision and hearing. Current research reports that CaBP4 gene mutations are mainly related to type 2B CSNB. In a study on CaBP4 in the retina, CaBP4 is co-localized with the Cav1.4 VDCC, and CaBP4 regulates its function through the IQ domain of Cav1.4 (11). In a study of CaBP4 gene knockout mice, the photosensitive synapses in CaBP4 gene-deficient mice were severely damaged in both anatomy and function. In situ hybridization and confocal microscopy showed that CaBP4 was specifically located in the photoreceptor and mainly located at the end of its synapse. Light and electron microscopy showed significant changes in photosensitive synapses, including a thinner outer plexiform layer and a decrease in the number of synapses and photosensitive terminals. The lack of CaBP4 also leads to the formation of ectopic synapses between the rod and rod bipolar or horizontal cells in the outer nuclear layer (11).
In the present study, human neuron cells transfected with the mutant recombinant plasmid (pEGFP-N1-CaBP4-p.G155D) constituted the mutation experimental group, human neuron cells transfected with the wild recombinant plasmid (pEGFP-N1-CaBP4) constituted the wild control group, and human neuron cells transfected with the blank recombinant plasmid (pEGFP-N1) constituted the blank control group. By using in vitro cell transfection, fluorescence microscopy showed that under the premise of applying the same transfection reagent and dosage and the same transfection plasmid dosage, the number of neuronal cells in the blank control group was higher than that in the mutation experiment group or the wild control group. From this, it can be understood that the CaBP4 protein with normal function in an over-expressed state has neuronal cytotoxicity. The cell death rate of the mutation experimental group was similar to that of the blank control group but higher than that of the wild control group. Combined with the normal function of the CaBP4 protein obtained above, it demonstrates neurocytotoxicity. It can be assumed that the function of the mutant CaBP4 protein is better than that of the wild type, which is weaker. Real-time fluorescent PCR detection showed that the mRNA expression of the mutant experimental group was higher than that of the wild control group, indicating that the CaBP4 gene p.G155D mutation significantly increased the mRNA expression of CaBP4; in the subsequent western blot experiment, protein expression was compared. We found that the amount of CaBP4 protein expressed in the mutant experimental group was lower than that in the wild control group. Combining the above two experimental results, we found that when the mRNA expression of the mutation experiment group increased, the expression of CaBP4 protein was lower than that of the wild control group, indicating that the CaBP4 protein of the mutation experiment group was more prone to hydrolysis. Therefore, we speculate that the CaBP4 gene p.G155D mutation reduces the protein stability of CaBP4.
In conclusion, the CaBP4 gene p.G155D mutation weakens the function of the CaBP4 protein and reduces its stability, thereby affecting its regulation of neuronal L-type calcium channels and causing changes in neuronal cell excitability. Therefore, it is reasonable to infer that the CaBP4 gene is likely to be a new pathogenic gene in the Chinese ADNFLE population.
Acknowledgments
We express special thanks to Guangdong Provincial People’s Hospital for providing us with experimental guidance.
Funding: This study was supported by the National Natural Science Foundation of China (2018; No. 81701284), the Science and Technology Foundation of Guangzhou (2020; No. 202002030428), and the Natural Science Foundation of Guangdong Province (2019; No. 2019A1515011864).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-22-54/rc
Data Sharing Statement: Available at https://tp.amegroups.com/article/view/10.21037/tp-22-54/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-22-54/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kurahashi H, Hirose S. Autosomal Dominant Nocturnal Frontal Lobe Epilepsy. Created: May 16, 2002; Updated: March 15, 2018. Seattle (WA): University of Washington, 1993–2021.
- Chen ZH, Wang C, Zhuo MQ, et al. Exome sequencing identified a novel missense mutation c.464G>A (p.G155D) in Ca2+-binding protein 4 (CABP4) in a Chinese pedigree with autosomal dominant nocturnal frontal lobe epilepsy. Oncotarget 2017;8:78940-7. [Crossref] [PubMed]
- Scheffer IE, Bhatia KP, Lopes-Cendes I, et al. Autosomal dominant frontal epilepsy misdiagnosed as sleep disorder. Lancet 1994;343:515-7. [Crossref] [PubMed]
- Picard F, Bertrand S, Steinlein OK, et al. Mutated nicotinic receptors responsible for autosomal dominant nocturnal frontal lobe epilepsy are more sensitive to carbamazepine. Epilepsia 1999;40:1198-209. [Crossref] [PubMed]
- Yoshimura R, Yanagihara N, Terao T, et al. Inhibition by carbamazepine of various ion channels-mediated catecholamine secretion in cultured bovine adrenal medullary cells. Naunyn Schmiedebergs Arch Pharmacol 1995;352:297-303. [Crossref] [PubMed]
- Ferini-Strambi L, Sansoni V, Combi R. Nocturnal frontal lobe epilepsy and the acetylcholine receptor. Neurologist 2012;18:343-9. [Crossref] [PubMed]
- Becchetti A, Aracri P, Meneghini S, et al. The role of nicotinic acetylcholine receptors in autosomal dominant nocturnal frontal lobe epilepsy. Front Physiol 2015;6:22. [Crossref] [PubMed]
- Milligan CJ, Li M, Gazina EV, et al. KCNT1 gain of function in 2 epilepsy phenotypes is reversed by quinidine. Ann Neurol 2014;75:581-90. [Crossref] [PubMed]
- Picard F, Makrythanasis P, Navarro V, et al. DEPDC5 mutations in families presenting as autosomal dominant nocturnal frontal lobe epilepsy. Neurology 2014;82:2101-6. [Crossref] [PubMed]
- Combi R, Dalprà L, Ferini-Strambi L, et al. Frontal lobe epilepsy and mutations of the corticotropin-releasing hormone gene. Ann Neurol 2005;58:899-904. [Crossref] [PubMed]
- Haeseleer F, Imanishi Y, Maeda T, et al. Essential role of Ca2+-binding protein 4, a Cav1.4 channel regulator, in photoreceptor synaptic function. Nat Neurosci 2004;7:1079-87. [Crossref] [PubMed]
- Park S, Li C, Haeseleer F, et al. Structural insights into activation of the retinal L-type Ca2+ channel (Cav1.4) by Ca2+-binding protein 4 (CaBP4). J Biol Chem 2014;289:31262-73. [Crossref] [PubMed]
- Shaltiel L, Paparizos C, Fenske S, et al. Complex regulation of voltage-dependent activation and inactivation properties of retinal voltage-gated Cav1.4 L-type Ca2+ channels by Ca2+-binding protein 4 (CaBP4). J Biol Chem 2012;287:36312-21. [Crossref] [PubMed]
- Xiong M, Su H, Xiang M. Research progress on pathogenesis of epilepsy. China Modern Medicine 2019;26:24-27.
- Beghi E. The Epidemiology of Epilepsy. Neuroepidemiology 2020;54:185-91. [Crossref] [PubMed]
- Steinlein OK. Genetic heterogeneity in familial nocturnal frontal lobe epilepsy. Prog Brain Res 2014;213:1-15. [Crossref] [PubMed]
- Nielsen TØ, Herlin MK, Linnet KM, et al. Autosomal dominant sleep-related hypermotor epilepsy caused by a previously unreported CHRNA4 variant. Eur J Med Genet 2022;65:104444. [Crossref] [PubMed]
- Zhuo M. The electrophysiologic function of CaBP4 geBe mutation in autosomal dominant noctumal frontal lobe epilepsy[D]. Guangzhou: Southern Medical University, 2015.
(English Language Editor: D. Fitzgerald)


