
Genetics of congenital heart disease: a narrative review of recent advances and clinical implications
Introduction
Congenital heart disease (CHD) is the most common birth defect, affecting nearly 1% of all live births (1). CHD encompasses a wide spectrum of defects from simple malformations with a favorable prognosis to more complex and severe lesions that require multiple catheter-based or surgical interventions with uncertain long-term outcomes. Although CHD remains a leading cause of morbidity and mortality in childhood, the population of adults with CHD is dramatically expanding. Now, more than 90% of children with CHD survive into adulthood due to significant advances in disease recognition and improved medical and surgical management across the lifespan (2-5). Therefore, understanding the genomic architecture of CHD is increasingly clinically important (6). While there have been significant advances in the elucidation of the genetic etiologies for other forms of inherited cardiac disease such as cardiomyopathy and arrhythmias, it has only been with the increased understanding of the molecular pathways regulating cardiovascular development over the past couple of decades that the genetic basis of CHD has become more defined (7-10). However, the detailed genetic architecture of CHD and how disruption of these underlying regulatory mechanisms result in the spectrum of CHD phenotypes is actively being investigated.
While numerous genes have been discovered to be implicated in the pathogenesis of syndromic CHD, the identification of the genetic contributors of non-syndromic CHD is more challenging due to genetic heterogeneity, incomplete segregation and potentially oligogenic or polygenic origins. The initial discoveries of disease-causing genes were primarily restricted to milder forms of CHD in non-syndromic and syndromic cases by using linkage analysis to study large families with autosomal dominant disease or by targeted sequencing of candidate genes in affected populations (7,8,10). Remarkable advances in genetic sequencing technologies, such as massively parallel or next-generation sequencing (NGS) have enabled the discovery of rare variants in new candidate genes that are likely contributing to non-syndromic CHD (11-15). Although in vivo and in vitro genetic models have allowed for assessment of the potential functional deficits of specific variants on gene function, these sequencing studies still have practical challenges in establishing pathogenicity of identified variants (16-18). Recent advances in powerful new technologies known as single-cell RNA sequencing (scRNA-seq) have facilitated the discovery of the role of individual cells during cardiac development and pathogenic mechanisms by which small subset of cells affected by genetic mutations lead to cardiac malformations (19).
In this review, we aimed to summarize the well-established genetic contributors to CHD and also discuss the recent advances in our understanding of the genetic architecture of CHD along with the challenges associated with the interpretation of newly discovered genetic variants in individuals with CHD. In addition, we sought to highlight the clinical implications of these genetic findings, which have the potential to predict and improve clinical outcomes in patients with CHD.
This narrative review was compiled through study, analysis, and discussion of previously published literature. PubMed was searched without time limitations and language restrictions, including articles related to the etiology and genetic contributors of CHD, NGS studies in large CHD cohorts, challenges with interpretation of NGS findings, functional genomics of CHD, and clinical implications of genetic testing and genetic prediction of clinical outcomes in patients with CHD. Search terms included congenital heart defects in combination with genetics, etiology, pathogenesis, mutations/genetic variation, environmental factors, NGS, exome sequencing, whole genome sequencing (WGS), variant prioritization, scRNA-seq, functional genomics, genetic animal models, human induced pluripotent stem cells (iPSCs), noncoding variants, or genetic testing and a combination of congenital heart disease, genetics and clinical outcomes. We present the following article in accordance with the Narrative Review reporting checklist (available at https://dx.doi.org/10.21037/tp-21-297).
Established etiologic contributors to CHD
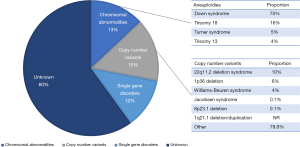
The etiology of CHD is multifactorial as both genetic and environmental factors have been implicated in its etiology (20). Specific genetic causes can be detected in an estimated 40% of CHD cases (Figure 1). Genetic causes of CHD are extremely heterogeneous, including chromosomal anomalies or aneuploidies (estimated 13%, range from 9% to 18%) (21), copy number variants (CNVs) (estimated 10–15%: range from 3% to 25% in syndromic CHD and 3% to 10% in non-syndromic CHD) (22-24), and single gene disorders (12%) (13,25-27). The genetic basis of CHD can be divided into syndromic CHD and non-syndromic CHD, where congenital abnormalities are isolated to the heart.
Genetic abnormalities associated with syndromic CHD
Numerous commonly observed syndromes have been found to be caused by chromosomal aneuploidies and CNVs as well as pathogenic variation of single genes (28). Common syndromes associated with CHD are summarized in Table 1. Chromosomal aneuploidies include the trisomies (13, 18 and 21) and monosomies such as Turner syndrome, that are detectable by karyotyping (29-34). CNVs are large deletions or duplications of DNA and pathogenic CNVs that are associated with syndromic CHD. These include 22q11.2 deletion syndrome (DiGeorge syndrome) (35-41), 1p36 deletion syndrome (42-44), 7q11.23 deletion (Williams-Beuren syndrome) (45,46), terminal deletions of 11q (Jacobsen syndrome) (47-50), 1q21.1 deletion/duplication (69-72), and 8p23.1 deletion syndrome (73,74), which can be detected by fluorescent in situ hybridization and/or chromosomal microarray (CMA). Syndromes caused by single gene variants have additionally been found to be genetically heterogeneous, including mutations in transcription factors and chromatin modifiers that are important for normal cardiac development. Single gene etiologies, inherited in a Mendelian manner, were initially detected by classic linkage analyses and targeted sequencing of candidate genes in large, multigenerational kindreds where multiple family members were affected with CHD associated with syndromes.
Table 1
| Syndrome | Gene | Loci | Cardiac defect | Clinical findings | References |
|---|---|---|---|---|---|
| Chromosomal aneuploidies | |||||
| Down syndrome | Unknown | Trisomy 21 | AVSD, ASD, VSD, PDA, TOF | CHD (40–50%), short stature, cognitive deficits, atlantoaxial instability, immune system dysfunction, hypotonia, hypothyroidism | ( |
| Turner syndrome | Unknown | 45, X (monosomy X) | CoA, BAV, dilated Ao, AS, HLHS | CHD (30%), short stature (partially growth hormone responsive), cognitive deficits, ADHD, lymphedema, webbed neck, primary amenorrhea | ( |
| CNVs | |||||
| 22q11.2 deletion syndrome (DiGeorge syndrome) |
|
22q11.2 deletion | Conotruncal defects (TOF, PTA), VSD, IAA, ASD | CHD (74–85%), cleft palate, bifid uvula, velopharyngeal insufficiency, microcephaly, hypocalcemia/hypoparathyroidism, thymic hypoplasia/immune deficit, psychiatric disorder, learning disability | ( |
| 1p36 deletion syndrome | Unknown | 1p36 deletion | PDA, VSD, ASD, BAV, Ebstein’s anomaly, LVNC | CHD (70%), Growth deficiency, intellectual disability, microcephaly, deep-set eyes, low-set ears, hearing loss, hypotonia, seizures, central nervous system defects, genital anomalies | ( |
| Williams-Beuren syndrome |
|
7q11.23 | SVAS, PPS, VSD, ASD | CHD (80%), dysmorphic facies, thick lips, strabismus, stellate iris pattern, intellectual disability, infantile hypercalcemia | ( |
| Jacobsen syndrome |
|
11q terminal deletion | HLHS, AS, VSD, CoA, Shone’s complex | CHD (56%), growth retardation, developmental delay, thrombocytopenia, platelet dysfunction, widely spaced eyes, strabismus, broad nasal bridge, thin upper lip, prominent forehead, intellectual disability | ( |
|
|
|||||
| Single-gene variation | |||||
| Alagille syndrome |
|
20p12.2 | PPS, TOF, PA | CHD (>90%), bile duct paucity, posterior embryotoxon, butterfly vertebrae, renal defects | ( |
|
|
1p12-p11 | ||||
| Char syndrome |
|
6p12.3 | PDA, VSD | CHD (58%), wide-set eyes, down-slanting palpebral fissures, thick lips, hand anomalies | ( |
| CHARGE syndrome |
|
8q12 | TOF, PDA, DORV, AVSD, VSD | CHD (75–85%), coloboma, choanal atresia, genital hypoplasia, ear anomalies, hearing loss, developmental delay, growth retardation, intellectual disability | ( |
| Costello syndrome |
|
11p15.5 | PS, ASD, VSD, HCM, arrhythmias | CHD (44–52%), short stature, feeding problems, broad facies, bitemporal narrowing, redundant skin, intellectual disability | ( |
| Ellis-van Creveld syndrome |
|
4p16.2 | Common atrium | CHD (60%), skeletal dysplasia, short limbs, polydactyly, short ribs, dysplastic nails, respiratory insufficiency | ( |
|
|
4p16.2 | ||||
| Holt-Oram syndrome |
|
12q24.1 | VSD, ASD, AVSD, conduction defects | CHD (50%), absent, hypoplastic, or triphalangeal thumbs, phocomelia, defects of radius, limb defects more prominent on left | ( |
| Kabuki syndrome |
|
12q13 | CoA, BAV, VSD, TOF, TGA, HLHS | CHD (50%), growth deficiency, wide palpebral fissures, large protuberant ears, fetal finger pads, intellectual disability, clinodactyly | ( |
|
|
Xp11.3 | ||||
| Noonan syndrome |
|
12q24.13 | Dysplastic PVS, ASD, TOF, AVSD, HCM, VSD, PDA | CHD (75%), short stature, hypertelorism, down-slanting palpebral fissures, ptosis, low posterior hairline, pectus deformity, bleeding disorder, chylothorax, cryptorchidism | ( |
|
|
2p22.1 | ||||
|
|
3p25.2 | ||||
|
|
12p12.1 | ||||
|
|
1p13.2 | ||||
|
|
1q22 | ||||
|
|
10q25.2 | ||||
|
|
14q21.3 | ||||
|
|
7q34 | ||||
ADHD, attention deficit/hyperactivity disorder; AS, aortic stenosis; ASD, atrial septal defect; AVSD, atrioventricular septal defect; BAV, bicuspid aortic valve; CHARGE, coloboma, heart defects, choanal atresia, retarded growth and development, genital anomalies, and ear anomalies; CHD, congenital heart disease; CNVs, copy number variants; CoA, coarctation of the aorta; dilated Ao, dilated ascending aorta; DORV, double-outlet right ventricle; HCM, hypertrophic cardiomyopathy; HLHS, hypoplastic left heart syndrome; IAA, interruption of aortic arch; LVNC, left ventricular noncompaction cardiomyopathy; PA, pulmonary atresia; PDA, patent ductus arteriosus; PPS, peripheral pulmonary stenosis; PS, pulmonary stenosis; PTA, persistent truncus arteriosus; PVS, pulmonary valve stenosis; SVAS, supravalvular aortic stenosis; TGA, transposition of great arteries; TOF, tetralogy of Fallot; VSD, ventricular septal defect.
Monogenic causes of non-syndromic CHD
Pathogenic variants that result in non-syndromic CHD can be broadly divided into transcription factors, cell signaling molecules and cardiac structural proteins (17,25). Monogenic causes of non-syndromic CHD with sufficient evidence are summarized in Table 2 and briefly described below. The expression and function of these factors are critical for cardiac progenitor lineages and the spatiotemporal regulation and formation of the complex three-dimensional heart structure.
Table 2
| Gene | Cardiovascular malformation | OMIM | References |
|---|---|---|---|
| Transcription factors | |||
| |
ASD, VSD | 602937 | ( |
| |
ASD, VSD, AVSD, PS, TOF | 600576 | ( |
| |
ASD, VSD, DORV, TOF, BAV | 611496 | ( |
| |
PTA, TOF | 601656 | ( |
| |
AVSD, DORV, HLHS, ASD, VSD | 602406 | ( |
| |
TOF, LVNC, VSD | 602407 | ( |
| |
Left-sided lesions | 601594 | ( |
| |
TGA | 608771 | ( |
| |
AVSD, AS, CoA, VSD, HLHS, TOF | 107773 | ( |
| |
ASD, atrioventricular conduction delay, TOF, HLHS, VSD | 600584 | ( |
| |
PTA | 611770 | ( |
| |
DORV, TOF, IAA, PTA, VSD | 602054 | ( |
| |
AVSD, TOF, BAV, CoA, ASD, VSD | 601620 | ( |
| |
ASD, VSD, MS, DCM | 606061 | ( |
| |
DORV | 600602 | ( |
| |
TA, AVSD | 600489 | ( |
| |
TOF, DORV | 603693 | ( |
| Cell signaling and adhesion proteins | |||
| |
AVSD | 102576 | ( |
| |
TGA, DORV | 605194 | ( |
| |
ASD, AVSD | 607170 | ( |
| |
TOF, TGA, VSD | 603621 | ( |
| |
ASD, DORV, TGA, TOF | 602880 | ( |
| |
HLHS, VSD, PA | 121014 | ( |
| |
AVSD | 604674 | ( |
| |
TOF, PS | 601920 | ( |
| |
TGA, DORV, TOF, VSD | 601265 | ( |
| |
BAV, AS, HLHS, TOF, PS, ASD, VSD, CoA, DORV | 190198 | ( |
| |
TAPVR | 173490 | ( |
| |
BAV, CoA, AS | 602931 | ( |
|
|
BAV, AS, TOF | 605101 | ( |
| |
TOF, PDA, AS, BAV, CoA, IAA, VSD | 192240 | ( |
| Structural proteins | |||
| |
ASD, HCM, DCM, LVNC | 102540 | ( |
| |
MVP | 603057 | ( |
| |
SVAS | 130160 | ( |
| |
ASD, HCM, DCM | 160710 | ( |
| |
Ebstein’s anomaly, LVNC, HCM, DCM | 160760 | ( |
| |
PDA, TAA | 160745 | ( |
AS, aortic stenosis; ASD, atrial septal defect; AVSD, atrioventricular septal defect; BAV, bicuspid aortic valve; CoA, coarctation of the aorta; DCM, dilated cardiomyopathy; DORV, double-outlet right ventricle; HCM, hypertrophic cardiomyopathy; HLHS, hypoplastic left heart syndrome; IAA, interruption of aortic arch; LVNC, left ventricular noncompaction cardiomyopathy; MS, mitral valve stenosis; MVP, mitral valve prolapse; PA, pulmonary atresia; PDA, patent ductus arteriosus; PPS, peripheral pulmonary stenosis; PS, pulmonary stenosis; PTA, persistent truncus arteriosus; SVAS, supravalvular aortic stenosis; TA, tricuspid atresia; TAA thoracic aortic aneurysm; TAPVR total anomalous pulmonary venous return; TGA, transposition of great arteries; TOF, tetralogy of Fallot; VSD, ventricular septal defect.
Transcription factors
Transcription factors involved in cardiac development have been identified by genetic studies with multiple animal model systems (152,153). NKX, GATA and T-box family members constitute the core regulatory network that is responsible for normal cardiogenesis and are causative genes in CHD (154).
Mutations in the homeobox transcription factor NKX2–5 were first reported as the cause of non-syndromic CHD by studying four kindreds with autosomal dominant disease (98). The common phenotype associated with NKX2–5 mutations is atrial septal defect (ASD) along with atrioventricular conduction abnormalities (99). NKX2–5 mutations have since been reported in a wide spectrum of CHD, including ventricular septal defect (VSD), tetralogy of Fallot (TOF), subvalvar aortic stenosis (AS), pulmonary atresia and hypoplastic left heart syndrome (HLHS), as well as atrioventricular conduction abnormalities, leading to complete heart block and sudden cardiac death (100-103). Previous studies demonstrated that mutations in the homeodomain of NKX2–5 are a cause of ASD, while mutations outside the homeodomain may result in TOF (155). In addition, mice harboring NKX2–5 mutations have been reported to recapitulate cardiac phenotypes found in humans (156-158). Identification of NKX2–5 mutations is clinically beneficial in terms of detecting patients with the increased risk of progressive conduction system disease, sudden cardiac death or asymptomatic ASD.
Mutations in the GATA family members GATA4, GATA5, and GATA6, characterized by zinc finger domains and transcriptional activation domains, have been identified in patients with various types of CHD. Heterozygous mutations in GATA4 were first discovered in familial cardiac septal defects (76). GATA4 mutations have been associated with ASD, VSD, atrioventricular septal defect (AVSD), pulmonary stenosis (PS), and TOF (77-80). These findings have been supported by the reports of similar cardiac phenotypes in mice haploinsufficient for GATA4 or those harboring disease-causing GATA4 mutations (159-161). Additionally, rare sequence variants in GATA5 have been reported in affected individuals with CHD, including bicuspid aortic valve (BAV), VSD, TOF and double outlet right ventricle (DORV) (82,162,163). Genetic deletion of GATA5 and endothelial cell-specific deletion of GATA5 using Tie2-Cre in mice led to BAV (164). Furthermore, GATA6 mutations were first reported in patients with persistent truncus arteriosus (PTA) (85) and have been implicated in a variety of CHD, including TOF, DORV, transposition of the great arteries (TGA), ASD, VSD and PS (86,165). However, murine models display BAV, including mice haploinsufficient for GATA6 and those with second heart field specific deletion of GATA6 using Isl1-Cre (166). Interestingly, GATA6 mutations were also found to be an important cause of pancreatic abnormalities (hypoplasia and agenesis) and associated
The T-box family consists of important transcription factors in cardiac development. TBX5 and TBX1 are implicated in the etiology of Holt-Oram syndrome and 22q11.2 deletion syndrome, respectively (168,169). In addition to the link to syndromic CHD, mutations in TBX5 and TBX1 have been identified in non-syndromic CHD such as TOF and cardiac septal defects (106,107,170). Another member of the family, TBX20, has subsequently been implicated in non-syndromic CHD, including cardiac septal defects, mitral valve stenosis, dilated cardiomyopathy and TOF (108-110).
Cell signaling and adhesion molecules
Notch signaling is important for cellular differentiation regulating the development of cardiac valves and chambers, and is associated with syndromic as well as non-syndromic CHD (51,129,171). Variants in NOTCH1 were the first reported genetic cause of aortic valve disease (133) and then have been described in not only left-sided CHD such as BAV, AS, coarctation of the aorta (CoA), and HLHS, but also TOF and other right-sided CHD (15,134,172).
Another important cell signaling pathway in cardiovascular development is the Nodal signaling that regulates left-right patterning. NODAL mutations have been reported in patients with heterotaxy as well as non-syndromic CHD including TGA, conotruncal heart defects and VSD (123,132). Mutations in several downstream targets of NODAL (GDF1, CFC1 and FOXH1) were also identified in CHD cohorts (118,124,173,174).
Structural proteins
Cardiac sarcomere and extracellular matrix proteins are crucial for the structure and function of cardiac muscle. Mutations in structural cardiac proteins are common causes of cardiomyopathy; however, some of these genes have also been associated with non-syndromic CHD. MYH6 (α-myosin heavy chain 6) mutations have been described in familial ASD along with hypertrophic or dilated forms of cardiomyopathy (146,175). Mutations in MYH7 (β-myosin heavy chain) have been associated with Ebstein’s anomaly and left ventricular noncompaction (LVNC) (149,150). Similarly, mutations in ACTC1 (α-cardiac actin), another sarcomere protein gene, have been identified in familial ASD and cardiomyopathy (140,176). MYH11 (myosin heavy chain 11) mutations have been implicated in familial thoracic aortic aneurysm with patent ductus arteriosus (PDA) (11,151). ELN (elastin) haploinsufficiency causes syndromic CHD in Williams-Beuren syndrome (45,142), whereas mutations in ELN have been reported in non-syndromic supravalvular AS and PS (143,144).
Environmental contributors to CHD
Environmental causes are implicated in 2–10% of CHD cases, and include maternal illnesses such as diabetes mellitus, obesity and phenylketonuria, maternal infection such as rubella and influenza, nutritional deficiencies such as folic acid, vitamin A and vitamin D, and teratogens such as thalidomide, alcohol, smoking and drugs (177-181). Although a significant proportion of CHD cases are likely to have some environmental etiologic contribution, it has been difficult to quantify the specific role these environmental contributors play in disease development. The underlying mechanisms by which environmental factors disrupt molecular pathways during cardiac development to cause CHD remain unknown. Moreover, CHD has been shown to be caused by gene-environment interactions in mice, where haploinsufficiency of Notch1 in developing embryos together with maternal hyperglycemic or hypoxic exposure resulted in increased incidence of CHD (182-184).
Recent advances in the understanding of the genetic architecture of CHD
Summary of NGS in large CHD cohorts
Over the past decade, remarkable advancements in NGS technologies have allowed for the identification of novel genetic etiologies for CHD and better understanding of the complex genetic architecture of non-syndromic CHD. Recent multi-institutional studies describing the results of exome sequencing of 2,871 CHD probands (among which were 2,645 trios) conducted by the Pediatric Cardiac Genomics Consortium (PCGC) have uncovered damaging rare transmitted variants and de novo variants (DNVs) in 8% of patients with sporadic CHD including 28% of syndromic and 3% of non-syndromic CHD (14,185). These DNVs were frequent in genes associated with proteins that function in chromatin modification, transcriptional regulation and RNA processing. A recent gene-burden analysis of 2,391 CHD trios revealed that cilia-related genes are enriched for rare, damaging recessive variants but comparatively less enriched for damaging DNVs (186). In contrast, chromatin-modifying genes were highly enriched for damaging DNVs. Similar findings were observed in another exome sequencing study that confirmed an excess of DNVs in chromatin-modifying genes involved in H3K4 methylation, H3K27 methylation, and H2BK120 ubiquitination (25). These DNVs in chromatin-modification genes were also associated with neurodevelopmental delays and extra-cardiac anomalies, indicating a potential role of these DNVs in syndromic forms of CHD. Furthermore, a large international study using exome sequencing of 1,891 probands has revealed a distinct genetic architecture for syndromic versus non-syndromic CHD, with unique enrichment of loss-of-function DNVs in syndromic CHD and incompletely penetrant inherited protein-truncating variants in non-syndromic CHD, respectively (13). Incomplete penetrance of these rare variants may contribute to the phenotypic heterogeneity in familial CHD and could also contribute to oligogenic causes of CHD (187,188).
With the advances in NGS technologies and large patient cohort studies, new genes and genetic variants were implicated in the pathogenesis of CHD as well as identifying a subset of patients with pathogenic variants identified in known CHD genes as described in Table 2. Variants in the gene, FLT4, which encodes vascular endothelial growth factor receptor 3 (VEGFR3), were identified in several large cohort exome sequencing studies. Jin et al. (14) found 2.3% of TOF patients to have LOF FLT4 variants and Page et al. (15) reported that deleterious FLT4 variants were identified in 2.4% of TOF cases. Majority of FLT4 variants were predicted LOF variants. A recent study of WGS in 231 individuals with CHD, most with TOF, demonstrated a significant truncating variant burden for FLT4 (189). Not surprisingly, these studies also identified pathogenic variants in NOTCH1 in patients with TOF. Additional studies are necessary to understand the mechanistic roles of FLT4 variants in the etiology of TOF, which may influence the subtypes of TOF and clinical outcomes (190). In addition to FLT4, other genes in the vascular endothelial growth factor (VEGF) pathway have been associated with TOF. LOF variants in KDR, encoding the vascular endothelial growth factor receptor 2 (VEGFR2), were identified in TOF cohorts (138) and more recently, exome sequencing in a familial case of TOF and large-scale genetic studies revealed rare variants in KDR in a family with TOF (191). These studies proposed novel mechanisms that dysregulated VEGF signaling contributes to the pathogenesis of TOF.
Recessive genotypes in MYH6 were identified ~11% of patients with Shone complex, and left ventricular dysfunction was demonstrated in 4 of 7 probands with MYH6 mutations in the recent large exome sequencing study by PCGC (14). A study of WGS in 5 patients with HLHS and reduced right ventricular ejection fraction revealed recessive compound heterozygous MYH6 variants in 2 patients (192). Furthermore, in a case-control study in 190 unrelated patients with HLHS, an increased burden of damaging MYH6 variants (10.5%) was noted compared with controls (2.9%) (193). A recent WGS study in 197 probands with HLHS identified rare, damaging MYH6 variants in 10% of cohorts (194). Interestingly, these studies suggest that MYH6 variants are associated with poor clinical outcomes in HLHS, as will be discussed further in the section titled genetic prediction of clinical outcomes.
RBFOX2, an RNA-binding protein that regulates alternative splicing, has also been identified as a new candidate CHD gene (195). RBFOX2 has a variety of biological activity contributing to neuronal maturation and axon assembly as well as cardiac remodeling and cardiac decompensation in pressure overload-induced heart failure (196,197). Damaging DNVs in RBFOX2 have been found in patients with HLHS in the large patient cohort studies (14,27). In addition, Verma et al. (198) uncovered a new role of RBFOX2 in the pathogenesis of HLHS, demonstrating that RBFOX2 was functionally impaired in HLHS patients leading to transcriptomic changes in the right ventricle of HLHS patients.
KMT2D encodes the histone-lysine N-methyltransferase 2D enzyme, responsible for H3K4 methylation, and regulates the genes involved in early embryonic development (199). Pathogenic variants in KMT2D are a well-known genetic cause of Kabuki syndrome (200). Recently, significant enrichment of damaging DNVs in KMT2D have been observed in CHD cases (14,27), and clustering of missense KMT2D variants have been found to cause a novel phenotype distinct from Kabuki syndrome, that includes CHD (201).
Challenges with interpretation of NGS results
Although NGS technologies in CHD cohorts have contributed to identifying variants associated with CHD risk, NGS techniques have some challenges that have limited translation to the clinical setting. First, there are difficulties in variant prioritization and interpretation. Second, establishing the pathogenicity of identified variants is still challenging even with advances in in vitro and in vivo genetic modeling of CHD. Third, with the increased availability and use of WGS, the role of noncoding variation in the genetic architecture of CHD is not clear.
Variant prioritization
In spite of the tremendous advances in genetic sequencing technologies, the identification of pathogenic variants in patients with non-syndromic CHD has been challenging, even in familial cases of CHD. Previously, we and others have successfully used exome sequencing and a variant prioritization pipeline to identify novel pathogenic variants in familial CHD using known CHD-causing gene lists. Blue et al. (202) reported that a targeted NGS gene panel of 57 candidate genes in a cohort of CHD families identified pathogenic variants in 31% of families. Consequently, they have successfully performed exome sequencing along with bioinformatics pipelines and filtering strategies to identify candidate variants in familial non-syndromic CHD (203). In our work, we identified a genetic etiology in 3/9 (33%) of familial cases of CHD using similar approaches (11). In these studies, variants were filtered using bioinformatics pipelines and prioritized according to in silico prediction programs, predicted mode of inheritance, minor allele frequencies, and presence in databases such as dbSNP (Single Nucleotide Polymorphism Database), the National Heart, Lung and Blood Institute Exome Sequencing Projects (ESP) (https://evs.gs.washington.edu/EVS/), 1000 Genomes (https://www.internationalgenome.org/), the Exome Aggregation Consortium (ExAC) and the Genome Aggregation Database (gnomAD) (https://gnomad.broadinstitute.org/). Variants were then classified based on the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP) guidelines (204). Since the release of these guidelines, several online tools and repositories have been developed for classification and interpretation of genetic variants, including the National Institutes of Health-funded Clinical Genome Resource (ClinGen) (https://clinicalgenome.org/working-groups/sequence-variant-interpretation/), Varsome (https://varsome.com/), Franklin (https://franklin.genoox.com/clinical-db/home) and Clinvar (https://www.ncbi.nlm.nih.gov/clinvar/). Recently, Szot et al. (205) reported the utility of their dual approaches for analyzing exome sequencing data to identify likely pathogenic variants. They achieved overall high diagnostic rate in families with sporadic and familial CHD by interrogating high confidence CHD-causing genes as well as an unbiased screen in which the exome sequencing data were analyzed comprehensively for additional variants not identified through the CHD gene list. In addition, a recent study has reported the effectiveness of pathway enrichment analyses of DNVs in exomes of CHD patients to explore novel CHD risk genes and validate potentially damaging variants (206).
With the advent of scRNA-seq technology, the profiling and analysis of single-cell transcriptomes is now possible with unprecedented resolution and throughput. The resolution of scRNA-seq datasets coupled with machine learning approaches has led to numerous findings in CHD. scRNA-seq has been utilized to generate cell atlases of cardiac cell types at various stage of cardiac development by profiling the anatomical locations of the embryonic heart (207,208). Additionally, scRNA-seq has been used to elucidate the mechanisms regulating the emergence and segregation of the early cardiac lineage during heart development (209,210). A network-based computational method for scRNA-seq analysis has revealed the mechanisms by which relatively small populations of cells are affected during cardiogenesis and how regulatory defects in discrete cell subsets can lead to morphologic developmental defects (19). Single-cell genomics will help to understand the pathogenic mechanisms of CHD at the single-cell level and provide novel insights into the genetic architecture of CHD. These approaches can refine variant prioritization pipeline by filtering novel candidate genes from large patient cohorts along with validation of cardiac expression from scRNA-seq datasets to expand our ability to identify pathogenic variants in CHD. The incorporation of these additional filtering criteria will result in continued improvement of the pathogenic variant detection algorithm and increasingly identify the genetic underpinnings of CHD.
Functional genomics of CHD
Although the ability to identify potential genetic variants contributing to CHD is improving, the determination of variant pathogenicity remains challenging. Accordingly, genetic model systems are required to characterize potentially pathogenic variants and elucidate pathogenic mechanisms (18). Numerous in vivo and in vitro models are available, including zebrafish, fruit fly, frog, chick and large mammals, and each model has strengths and weaknesses. Murine models have been widely used to study cardiovascular development because they share a high degree of sequence conservation to humans and recapitulate human cardiac development. However, genotype-phenotype relationships can be different between humans and mice. Furthermore, using the murine model system is not amenable for the analysis of large number of recently identified genetic variants. Therefore, additional genetic models that allow for a higher throughput analysis are increasingly necessary.
Human iPSCs provide a unique in vitro platform to study genetic mechanisms of non-syndromic CHD by single-gene defects (211). Human iPSCs can be differentiated into a variety of cell types including cardiomyocytes (iPSC-CMs), endothelial cells (iPSC-ECs), vascular smooth muscle cells (iPSC-SMCs) and cardiac fibroblasts (iPSC-CFs). In addition, patient-specific iPSCs can be tailored to the unique individual genetics of patients and are crucial in investigating the complex genetic mechanisms of CHD by incorporating with advanced NGS technologies as well as transgenic animal models. These significant advantages have made it possible to expand the use of patient-derived iPSCs to study a variety of CHD, including supravalvular AS (212), and BAV and calcific aortic valve disease (213), cardiac septal defects (214,215), HLHS (216-219), pulmonary atresia with intact ventricular septum (220), and LVNC (187,221). A recent examination of transgenic murine hearts and patient-derived iPSC-CMs study revealed the oligogenic inheritance of LVNC with the evidence for a NKX2–5 variant as a genetic modifier (187). Another recent study reported genome-wide transcriptome profiles of iPSC-CMs that were generated from patients with single ventricle heart disease and non-syndromic TOF (222). These studies provide growing evidence for the effectiveness of using iPSCs to model CHD and open the door to identifying how a modifier gene or multiple genes interact to cause CHD. Furthermore, the single-cell omics approaches in cardiovascular precision medicine combined with iPSC platforms, epigenomics and proteomics will contribute to the development of patient-specific therapeutics for CHD.
Noncoding genetic variation
WGS captures both coding and noncoding regions of the genome. Although the overall yield of exome sequencing in patients with CHD is low, WGS allows for the discoveries of a large number of CNVs and single nucleotide variations (SNVs) in regulatory and noncoding regions of the entire human genome. However, establishing a link between noncoding genetic variation and CHD is still challenging because classic transgenic mouse methods are not as applicable to noncoding regions. Allele-specific expression analysis can identify candidate noncoding genetic variants by combining RNA-seq and WGS in complex genetic diseases including CHD (195,223). The largest WGS in CHD demonstrated an enrichment in damaging DNVs in noncoding regions in CHD trios compared to controls and estimated that noncoding DNVs are associated with 17–45% of CHD cases (224). These studies highlight the potential of WGS to elucidate the role of noncoding variants contributing to the pathogenesis of CHD. Future studies are needed to establish the transcriptional and post-transcriptional regulatory effects of noncoding variants on cardiac development.
Clinical implications of recent genetic advances
Genetic testing
Over the past 20 years, the advanced genetic testing methodologies (e.g., CMA and exome sequencing) are increasingly being incorporated into the genetic evaluation of patients with CHD, and the results of this testing has important clinical implications. The clinical benefits of genetic testing for patients with CHD include establishing a genetic diagnosis, anticipatory management of CHD and associated extra-cardiac conditions, and clinical screening of at-risk family members (225). In addition, genetic testing can provide the information about the genetic causes and the recurrence risk of CHD to support reproductive decisions and to guide perinatal management. While CMA and single-gene testing for specific syndromes are currently utilized as standard genetic testing for patients with CHD and extra-cardiac abnormalities, diagnostic use of NGS technology is evolving, allowing the interrogation of large datasets of genetic variants. The overall diagnostic yield of genetic testing varies from the low single digits to close to 40%, which depends on the tests available, the anatomical lesions, and the presence of extra-cardiac or other relevant clinical features (13,27). Despite the potential utility, variant interpretation is challenging as genotype-phenotype correlations remain elusive and phenotypic heterogeneity or incomplete penetrance is present. Communicating complex genetic findings to potentially unsuspecting patients is also challenging. Given the potential psychosocial impact of genetic testing in asymptomatic, phenotype-negative individuals, it is therefore critical that genetic testing should be offered within the context of appropriate genetic counseling so that families are given the opportunity to discuss insurance and other risks well in advance (6).
Genetic prediction of clinical outcomes
Despite advances in our understanding of genetics underlying CHD, prediction of clinical outcomes by using genetic findings remains challenging. Initial successful examples of linking common genetic variants for clinical care were limited to areas of prediction of disease risk, disease classification, and drug response (226). Apolipoprotein E genotypes were associated with adverse neurodevelopmental outcomes after cardiac surgery in patients with CHD (227,228). Additionally, in patients with single ventricle, adrenergic receptor genotypes were associated with poor postoperative outcomes, and renin-angiotensin-aldosterone receptor genotypes were linked to failure of ventricular remodeling after surgery, impaired renal function and somatic growth (229).
Discovery of an increasing number of genetic contributors to CHD, large-scale data of clinical phenotypes and the growing knowledge about the biological and physiological impact of genetic variants are providing evidence to help understand clinical implications of genetic findings to predict clinical outcomes in CHD patients. Underlying genetic variants are increasingly recognized to affect clinical outcomes such as long-term/event-free survival, growth, neurodevelopmental performance, and ventricular function (230). Additionally, a significant increase in genetic burden of novel and rare variants in genes implicated in CHD and neurodevelopmental disabilities (NDD) was identified to be associated with the development of NDD in CHD patients (231). Therefore, identification of genetic variants associated with CHD and neurodevelopmental abnormalities could be useful for early learning intervention strategies.
Furthermore, specific genetic variation influences clinical outcomes in some categories of CHD patients. Pathogenic variants in cilia genes may predict postoperative and respiratory outcomes (25). Patients with single ventricle heart defects segregated by the presence of potentially pathogenic CNVs had worse outcomes as well as worse linear growth and neurodevelopmental performance at 14 months of age than those without CNVs (22). Pathogenic CNVs have been associated with increased risk of death or transplant in non-syndromic CHD (23). A 22q11.2 deletion in CHD patients also affects their surgical complication rate and survival. Patients with TOF and 22q11.2 deletion syndrome were found to have a longer stay in the intensive care unit and a worse quality of life (232). A recent study of exome sequencing in a large cohort of 2,517 patients with CHD demonstrated that 11.7% of patients carried clinically significant DNVs and patients with DNVs were more likely to have extra-cardiac anomalies (233). This study also found that DNVs were associated with lower transplant-free survival and worse postoperative respiratory outcomes such as longer times on the ventilator in patients who underwent open-heart surgery. Interestingly, the magnitude of the association between DNVs and clinical outcomes was shown to be different for patients with versus without extra-cardiac anomalies. In patients with extra-cardiac anomalies, the association of DNVs with worse outcomes was modest without statistical significance. In contrast, DNVs were strongly associated with adverse outcomes among patients without extra-cardiac anomalies. These important findings suggest a benefit for genetic testing even in patients without extra-cardiac anomalies who are not suspected to have genetic abnormalities in routine clinical practice.
Although only a few HLHS candidate genes have been validated by robust genetic and functional studies, several genetic factors have been reported to have an impact on clinical outcomes in patients with HLHS. Patients with HLHS harboring pathogenic CNVs were reported to be associated with significantly decreased survival compared to those with normal CMA or variants of undetermined significance (VUS) (234). In addition, HLHS with risk of poor outcomes has been linked to MYH6 variants. Patients harboring MYH6 variants had abnormal myocardial physiology and reduced right ventricular ejection fraction (192). Transplant-free survival was significantly lower in HLHS patients with damaging MYH6 variants compared with HLHS patients without MYH6 variants (193). Most recently, compound heterozygosity for rare damaging variants in MYH6 or MYBPC3, encoding myosin binding protein C3, was found to be a risk factor for myocardial dysfunction in patients with HLHS (194). These findings could help identify patients at risk for poor outcomes and develop precision medicine approaches tailored to the genetic information of each patient in the future.
Future directions
The increased understanding of genetics of CHD is providing new insights into the etiology of CHD as well as the impact of genetic variants on clinical outcomes in patients with CHD. More accurate detection and interpretation of pathologic genetic variants in CHD patients will enable clinicians to identify extra-cardiac manifestations, predict post-operative course and long-term outcomes, leading to the improvements of clinical outcomes. Further refinement of the clinical variant interpretation framework such as ACMG/AMP guidelines will construct a more accurate, consistent and transparent approach to variant classification. In addition, larger studies in well-phenotyped CHD cohorts, including important long-term clinical outcomes, will be required to determine further genetic factors contributing to the pathogenesis of CHD and its associated morbidities.
The ultimate goals are to develop therapies to slow the progression or prevent the occurrence of CHD. Current advances in sequencing technologies and functional genomic models of CHD will allow for the integration of genome editing, cardiac bioengineering and cardiac organoid models. The maturation of these technologies will open the door for new regenerative and preventive therapeutic approaches to treat the core disease mechanisms in CHD patients in the future.
Acknowledgments
Funding: JY was supported by Japan Heart Foundation/Bayer Yakuhin Research Grant Abroad and VG is supported by funding from NIH/NHLBI (R01-HL144009) and Additional Ventures Foundation.
Footnote
Reporting Checklist: The authors have completed the Narrative Review Checklist. Available at https://dx.doi.org/10.21037/tp-21-297
Peer Review File: Available at https://dx.doi.org/10.21037/tp-21-297
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tp-21-297). VG serves as an unpaid editorial board member of Translational Pediatrics from Aug 2021 to Jul 2023. The other author has no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hoffman JI, Kaplan S. The incidence of congenital heart disease. J Am Coll Cardiol 2002;39:1890-900. [Crossref] [PubMed]
- Boneva RS, Botto LD, Moore CA, et al. Mortality associated with congenital heart defects in the United States: trends and racial disparities, 1979-1997. Circulation 2001;103:2376-81. [Crossref] [PubMed]
- Gilboa SM, Salemi JL, Nembhard WN, et al. Mortality resulting from congenital heart disease among children and adults in the United States, 1999 to 2006. Circulation 2010;122:2254-63. [Crossref] [PubMed]
- Khairy P, Ionescu-Ittu R, Mackie AS, et al. Changing mortality in congenital heart disease. J Am Coll Cardiol 2010;56:1149-57. [Crossref] [PubMed]
- Raissadati A, Nieminen H, Jokinen E, et al. Progress in late results among pediatric cardiac surgery patients: a population-based 6-decade study with 98% follow-up. Circulation 2015;131:347-53; discussion 353. [Crossref] [PubMed]
- Mital S, Musunuru K, Garg V, et al. Enhancing Literacy in Cardiovascular Genetics: A Scientific Statement From the American Heart Association. Circ Cardiovasc Genet 2016;9:448-67. [Crossref] [PubMed]
- Garg V. Insights into the genetic basis of congenital heart disease. Cell Mol Life Sci 2006;63:1141-8. [Crossref] [PubMed]
- Richards AA, Garg V. Genetics of congenital heart disease. Curr Cardiol Rev 2010;6:91-7. [Crossref] [PubMed]
- Zaidi S, Brueckner M. Genetics and Genomics of Congenital Heart Disease. Circ Res 2017;120:923-40. [Crossref] [PubMed]
- Pierpont ME, Brueckner M, Chung WK, et al. Genetic Basis for Congenital Heart Disease: Revisited: A Scientific Statement From the American Heart Association. Circulation 2018;138:e653-711. [Crossref] [PubMed]
- LaHaye S, Corsmeier D, Basu M, et al. Utilization of Whole Exome Sequencing to Identify Causative Mutations in Familial Congenital Heart Disease. Circ Cardiovasc Genet 2016;9:320-9. [Crossref] [PubMed]
- Zahavich L, Bowdin S, Mital S. Use of Clinical Exome Sequencing in Isolated Congenital Heart Disease. Circ Cardiovasc Genet 2017;10:e001581 [Crossref] [PubMed]
- Sifrim A, Hitz MP, Wilsdon A, et al. Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nat Genet 2016;48:1060-5. [Crossref] [PubMed]
- Jin SC, Homsy J, Zaidi S, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet 2017;49:1593-601. [Crossref] [PubMed]
- Page DJ, Miossec MJ, Williams SG, et al. Whole Exome Sequencing Reveals the Major Genetic Contributors to Nonsyndromic Tetralogy of Fallot. Circ Res 2019;124:553-63. [Crossref] [PubMed]
- Garg V. Molecular genetics of aortic valve disease. Curr Opin Cardiol 2006;21:180-4. [Crossref] [PubMed]
- Fahed AC, Gelb BD, Seidman JG, et al. Genetics of congenital heart disease: the glass half empty. Circ Res 2013;112:707-20. [Crossref] [PubMed]
- Majumdar U, Yasuhara J, Garg V. In Vivo and In Vitro Genetic Models of Congenital Heart Disease. Cold Spring Harb Perspect Biol 2021;13:a036764 [Crossref] [PubMed]
- de Soysa TY, Ranade SS, Okawa S, et al. Single-cell analysis of cardiogenesis reveals basis for organ-level developmental defects. Nature 2019;572:120-4. [Crossref] [PubMed]
- Cowan JR, Ware SM. Genetics and genetic testing in congenital heart disease. Clin Perinatol 2015;42:373-93. ix. [Crossref] [PubMed]
- Hartman RJ, Rasmussen SA, Botto LD, et al. The contribution of chromosomal abnormalities to congenital heart defects: a population-based study. Pediatr Cardiol 2011;32:1147-57. [Crossref] [PubMed]
- Carey AS, Liang L, Edwards J, et al. Effect of copy number variants on outcomes for infants with single ventricle heart defects. Circ Cardiovasc Genet 2013;6:444-51. [Crossref] [PubMed]
- Kim DS, Kim JH, Burt AA, et al. Burden of potentially pathologic copy number variants is higher in children with isolated congenital heart disease and significantly impairs covariate-adjusted transplant-free survival. J Thorac Cardiovasc Surg 2016;151:1147-51.e4. [Crossref] [PubMed]
- Glessner JT, Bick AG, Ito K, et al. Increased frequency of de novo copy number variants in congenital heart disease by integrative analysis of single nucleotide polymorphism array and exome sequence data. Circ Res 2014;115:884-96. [Crossref] [PubMed]
- Zaidi S, Choi M, Wakimoto H, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature 2013;498:220-3. [Crossref] [PubMed]
- van der Bom T, Zomer AC, Zwinderman AH, et al. The changing epidemiology of congenital heart disease. Nat Rev Cardiol 2011;8:50-60. [Crossref] [PubMed]
- Homsy J, Zaidi S, Shen Y, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science 2015;350:1262-6. [Crossref] [PubMed]
- Muntean I, Togănel R, Benedek T. Genetics of Congenital Heart Disease: Past and Present. Biochem Genet 2017;55:105-23. [Crossref] [PubMed]
- Fudge JC Jr, Li S, Jaggers J, et al. Congenital heart surgery outcomes in Down syndrome: analysis of a national clinical database. Pediatrics 2010;126:315-22. [Crossref] [PubMed]
- Bull MJCommittee on Genetics. Health supervision for children with Down syndrome. Pediatrics 2011;128:393-406. [Crossref] [PubMed]
- Bull MJ. Down Syndrome. N Engl J Med 2020;382:2344-52. [Crossref] [PubMed]
- Sybert VP, McCauley E. Turner's syndrome. N Engl J Med 2004;351:1227-38. [Crossref] [PubMed]
- Gravholt CH, Andersen NH, Conway GS, et al. Clinical practice guidelines for the care of girls and women with Turner syndrome: proceedings from the 2016 Cincinnati International Turner Syndrome Meeting. Eur J Endocrinol 2017;177:G1-G70. [Crossref] [PubMed]
- Levitsky LL, Luria AH, Hayes FJ, et al. Turner syndrome: update on biology and management across the life span. Curr Opin Endocrinol Diabetes Obes 2015;22:65-72. [Crossref] [PubMed]
- Botto LD, May K, Fernhoff PM, et al. A population-based study of the 22q11.2 deletion: phenotype, incidence, and contribution to major birth defects in the population. Pediatrics 2003;112:101-7. [Crossref] [PubMed]
- Peyvandi S, Lupo PJ, Garbarini J, et al. 22q11.2 deletions in patients with conotruncal defects: data from 1,610 consecutive cases. Pediatr Cardiol 2013;34:1687-94. [Crossref] [PubMed]
- Mlynarski EE, Xie M, Taylor D, et al. Rare copy number variants and congenital heart defects in the 22q11.2 deletion syndrome. Hum Genet 2016;135:273-85. [Crossref] [PubMed]
- Digilio MC, Angioni A, De Santis M, et al. Spectrum of clinical variability in familial deletion 22q11.2: from full manifestation to extremely mild clinical anomalies. Clin Genet 2003;63:308-13. [Crossref] [PubMed]
- Marino B, Digilio MC, Toscano A, et al. Anatomic patterns of conotruncal defects associated with deletion 22q11. Genet Med 2001;3:45-8. [Crossref] [PubMed]
- Goldmuntz E, Clark BJ, Mitchell LE, et al. Frequency of 22q11 deletions in patients with conotruncal defects. J Am Coll Cardiol 1998;32:492-8. [Crossref] [PubMed]
- Momma K, Kondo C, Ando M, et al. Tetralogy of Fallot associated with chromosome 22q11 deletion. Am J Cardiol 1995;76:618-21. [Crossref] [PubMed]
- Battaglia A, Hoyme HE, Dallapiccola B, et al. Further delineation of deletion 1p36 syndrome in 60 patients: a recognizable phenotype and common cause of developmental delay and mental retardation. Pediatrics 2008;121:404-10. [Crossref] [PubMed]
- Heilstedt HA, Ballif BC, Howard LA, et al. Population data suggest that deletions of 1p36 are a relatively common chromosome abnormality. Clin Genet 2003;64:310-6. [Crossref] [PubMed]
- Slavotinek A, Shaffer LG, Shapira SK. Monosomy 1p36. J Med Genet 1999;36:657-63. [PubMed]
- Pober BR. Williams-Beuren syndrome. N Engl J Med 2010;362:239-52. [Crossref] [PubMed]
- Eronen M, Peippo M, Hiippala A, et al. Cardiovascular manifestations in 75 patients with Williams syndrome. J Med Genet 2002;39:554-8. [Crossref] [PubMed]
- Jacobsen P, Hauge M, Henningsen K, et al. An (11;21) translocation in four generations with chromosome 11 abnormalities in the offspring. A clinical, cytogenetical, and gene marker study. Hum Hered 1973;23:568-85. [Crossref] [PubMed]
- Grossfeld PD, Mattina T, Lai Z, et al. The 11q terminal deletion disorder: a prospective study of 110 cases. Am J Med Genet A 2004;129A:51-61. [Crossref] [PubMed]
- Favier R, Akshoomoff N, Mattson S, et al. Jacobsen syndrome: Advances in our knowledge of phenotype and genotype. Am J Med Genet C Semin Med Genet 2015;169:239-50. [Crossref] [PubMed]
- Ye M, Coldren C, Liang X, et al. Deletion of ETS-1, a gene in the Jacobsen syndrome critical region, causes ventricular septal defects and abnormal ventricular morphology in mice. Hum Mol Genet 2010;19:648-56. [Crossref] [PubMed]
- McDaniell R, Warthen DM, Sanchez-Lara PA, et al. NOTCH2 mutations cause Alagille syndrome, a heterogeneous disorder of the notch signaling pathway. Am J Hum Genet 2006;79:169-73. [Crossref] [PubMed]
- Turnpenny PD, Ellard S. Alagille syndrome: pathogenesis, diagnosis and management. Eur J Hum Genet 2012;20:251-7. [Crossref] [PubMed]
- McElhinney DB, Krantz ID, Bason L, et al. Analysis of cardiovascular phenotype and genotype-phenotype correlation in individuals with a JAG1 mutation and/or Alagille syndrome. Circulation 2002;106:2567-74. [Crossref] [PubMed]
- Satoda M, Zhao F, Diaz GA, et al. Mutations in TFAP2B cause Char syndrome, a familial form of patent ductus arteriosus. Nat Genet 2000;25:42-6. [Crossref] [PubMed]
- Trider CL, Arra-Robar A, van Ravenswaaij-Arts C, et al. Developing a CHARGE syndrome checklist: Health supervision across the lifespan (from head to toe). Am J Med Genet A 2017;173:684-91. [Crossref] [PubMed]
- Verloes A. Updated diagnostic criteria for CHARGE syndrome: a proposal. Am J Med Genet A 2005;133A:306-8. [Crossref] [PubMed]
- Lin AE, Alexander ME, Colan SD, et al. Clinical, pathological, and molecular analyses of cardiovascular abnormalities in Costello syndrome: a Ras/MAPK pathway syndrome. Am J Med Genet A 2011;155A:486-507. [Crossref] [PubMed]
- Ruiz-Perez VL, Ide SE, Strom TM, et al. Mutations in a new gene in Ellis-van Creveld syndrome and Weyers acrodental dysostosis. Nat Genet 2000;24:283-6. [Crossref] [PubMed]
- Ruiz-Perez VL, Tompson SW, Blair HJ, et al. Mutations in two nonhomologous genes in a head-to-head configuration cause Ellis-van Creveld syndrome. Am J Hum Genet 2003;72:728-32. [Crossref] [PubMed]
- Basson CT, Cowley GS, Solomon SD, et al. The clinical and genetic spectrum of the Holt-Oram syndrome (heart-hand syndrome) N Engl J Med 1994;330:885-91. [Crossref] [PubMed]
- McDermott DA, Bressan MC, He J, et al. TBX5 genetic testing validates strict clinical criteria for Holt-Oram syndrome. Pediatr Res 2005;58:981-6. [Crossref] [PubMed]
- Wessels MW, Brooks AS, Hoogeboom J, et al. Kabuki syndrome: a review study of three hundred patients. Clin Dysmorphol 2002;11:95-102. [Crossref] [PubMed]
- Hannibal MC, Buckingham KJ, Ng SB, et al. Spectrum of MLL2 (ALR) mutations in 110 cases of Kabuki syndrome. Am J Med Genet A 2011;155A:1511-6. [Crossref] [PubMed]
- Digilio MC, Marino B, Toscano A, et al. Congenital heart defects in Kabuki syndrome. Am J Med Genet 2001;100:269-74. [Crossref] [PubMed]
- Romano AA, Allanson JE, Dahlgren J, et al. Noonan syndrome: clinical features, diagnosis, and management guidelines. Pediatrics 2010;126:746-59. [Crossref] [PubMed]
- Tartaglia M, Mehler EL, Goldberg R, et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet 2001;29:465-8. [Crossref] [PubMed]
- Roberts AE, Allanson JE, Tartaglia M, et al. Noonan syndrome. Lancet 2013;381:333-42. [Crossref] [PubMed]
- Prendiville TW, Gauvreau K, Tworog-Dube E, et al. Cardiovascular disease in Noonan syndrome. Arch Dis Child 2014;99:629-34. [Crossref] [PubMed]
- Christiansen J, Dyck JD, Elyas BG, et al. Chromosome 1q21.1 contiguous gene deletion is associated with congenital heart disease. Circ Res 2004;94:1429-35. [Crossref] [PubMed]
- Digilio MC, Bernardini L, Consoli F, et al. Congenital heart defects in recurrent reciprocal 1q21.1 deletion and duplication syndromes: rare association with pulmonary valve stenosis. Eur J Med Genet 2013;56:144-9. [Crossref] [PubMed]
- Mefford HC, Sharp AJ, Baker C, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med 2008;359:1685-99. [Crossref] [PubMed]
- Guida V, Ferese R, Rocchetti M, et al. A variant in the carboxyl-terminus of connexin 40 alters GAP junctions and increases risk for tetralogy of Fallot. Eur J Hum Genet 2013;21:69-75. [Crossref] [PubMed]
- Claeys I, Holvoet M, Eyskens B, et al. A recognisable behavioural phenotype associated with terminal deletions of the short arm of chromosome 8. Am J Med Genet 1997;74:515-20. [Crossref] [PubMed]
- Wat MJ, Shchelochkov OA, Holder AM, et al. Chromosome 8p23.1 deletions as a cause of complex congenital heart defects and diaphragmatic hernia. Am J Med Genet A 2009;149A:1661-77. [Crossref] [PubMed]
- Sperling S, Grimm CH, Dunkel I, et al. Identification and functional analysis of CITED2 mutations in patients with congenital heart defects. Hum Mutat 2005;26:575-82. [Crossref] [PubMed]
- Garg V, Kathiriya IS, Barnes R, et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 2003;424:443-7. [Crossref] [PubMed]
- Okubo A, Miyoshi O, Baba K, et al. A novel GATA4 mutation completely segregated with atrial septal defect in a large Japanese family. J Med Genet 2004;41:e97 [Crossref] [PubMed]
- Hirayama-Yamada K, Kamisago M, Akimoto K, et al. Phenotypes with GATA4 or NKX2.5 mutations in familial atrial septal defect. Am J Med Genet A 2005;135:47-52. [Crossref] [PubMed]
- Sarkozy A, Conti E, Neri C, et al. Spectrum of atrial septal defects associated with mutations of NKX2.5 and GATA4 transcription factors. J Med Genet 2005;42:e16 [Crossref] [PubMed]
- Tomita-Mitchell A, Maslen CL, Morris CD, et al. GATA4 sequence variants in patients with congenital heart disease. J Med Genet 2007;44:779-83. [Crossref] [PubMed]
- Posch MG, Perrot A, Schmitt K, et al. Mutations in GATA4, NKX2.5, CRELD1, and BMP4 are infrequently found in patients with congenital cardiac septal defects. Am J Med Genet A 2008;146A:251-3. [Crossref] [PubMed]
- Jiang JQ, Li RG, Wang J, et al. Prevalence and spectrum of GATA5 mutations associated with congenital heart disease. Int J Cardiol 2013;165:570-3. [Crossref] [PubMed]
- Shi LM, Tao JW, Qiu XB, et al. GATA5 loss-of-function mutations associated with congenital bicuspid aortic valve. Int J Mol Med 2014;33:1219-26. [Crossref] [PubMed]
- Shan JP, Wang XL, Qiao YG, et al. Novel and functional DNA sequence variants within the GATA5 gene promoter in ventricular septal defects. World J Pediatr 2014;10:348-53. [Crossref] [PubMed]
- Kodo K, Nishizawa T, Furutani M, et al. GATA6 mutations cause human cardiac outflow tract defects by disrupting semaphorin-plexin signaling. Proc Natl Acad Sci U S A 2009;106:13933-8. [Crossref] [PubMed]
- Maitra M, Koenig SN, Srivastava D, et al. Identification of GATA6 sequence variants in patients with congenital heart defects. Pediatr Res 2010;68:281-5. [Crossref] [PubMed]
- Allen HL, Flanagan SE, Shaw-Smith C, et al. GATA6 haploinsufficiency causes pancreatic agenesis in humans. Nat Genet 2011;44:20-2. [Crossref] [PubMed]
- Lin X, Huo Z, Liu X, et al. A novel GATA6 mutation in patients with tetralogy of Fallot or atrial septal defect. J Hum Genet 2010;55:662-7. [Crossref] [PubMed]
- Yorifuji T, Kawakita R, Hosokawa Y, et al. Dominantly inherited diabetes mellitus caused by GATA6 haploinsufficiency: variable intrafamilial presentation. J Med Genet 2012;49:642-3. [Crossref] [PubMed]
- Reamon-Buettner SM, Ciribilli Y, Inga A, et al. A loss-of-function mutation in the binding domain of HAND1 predicts hypoplasia of the human hearts. Hum Mol Genet 2008;17:1397-405. [Crossref] [PubMed]
- Reamon-Buettner SM, Ciribilli Y, Traverso I, et al. A functional genetic study identifies HAND1 mutations in septation defects of the human heart. Hum Mol Genet 2009;18:3567-78. [Crossref] [PubMed]
- Shen L, Li XF, Shen AD, et al. Transcription factor HAND2 mutations in sporadic Chinese patients with congenital heart disease. Chin Med J (Engl) 2010;123:1623-7. [PubMed]
- Sun YM, Wang J, Qiu XB, et al. A HAND2 Loss-of-Function Mutation Causes Familial Ventricular Septal Defect and Pulmonary Stenosis. G3 (Bethesda) 2016;6:987-92. [Crossref] [PubMed]
- Töpf A, Griffin HR, Glen E, et al. Functionally significant, rare transcription factor variants in tetralogy of Fallot. PLoS One 2014;9:e95453 [Crossref] [PubMed]
- Li AH, Hanchard NA, Furthner D, et al. Whole exome sequencing in 342 congenital cardiac left sided lesion cases reveals extensive genetic heterogeneity and complex inheritance patterns. Genome Med 2017;9:95. [Crossref] [PubMed]
- Muncke N, Jung C, Rüdiger H, et al. Missense mutations and gene interruption in PROSIT240, a novel TRAP240-like gene, in patients with congenital heart defect (transposition of the great arteries). Circulation 2003;108:2843-50. [Crossref] [PubMed]
- Al Turki S, Manickaraj AK, Mercer CL, et al. Rare variants in NR2F2 cause congenital heart defects in humans. Am J Hum Genet 2014;94:574-85. [Crossref] [PubMed]
- Schott JJ, Benson DW, Basson CT, et al. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science 1998;281:108-11. [Crossref] [PubMed]
- Benson DW, Silberbach GM, Kavanaugh-McHugh A, et al. Mutations in the cardiac transcription factor NKX2.5 affect diverse cardiac developmental pathways. J Clin Invest 1999;104:1567-73. [Crossref] [PubMed]
- Goldmuntz E, Geiger E, Benson DW. NKX2.5 mutations in patients with tetralogy of fallot. Circulation 2001;104:2565-8. [Crossref] [PubMed]
- McElhinney DB, Geiger E, Blinder J, et al. NKX2.5 mutations in patients with congenital heart disease. J Am Coll Cardiol 2003;42:1650-5. [Crossref] [PubMed]
- Stallmeyer B, Fenge H, Nowak-Göttl U, et al. Mutational spectrum in the cardiac transcription factor gene NKX2.5 (CSX) associated with congenital heart disease. Clin Genet 2010;78:533-40. [Crossref] [PubMed]
- Ellesøe SG, Johansen MM, Bjerre JV, et al. Familial Atrial Septal Defect and Sudden Cardiac Death: Identification of a Novel NKX2-5 Mutation and a Review of the Literature. Congenit Heart Dis 2016;11:283-90. [Crossref] [PubMed]
- Heathcote K, Braybrook C, Abushaban L, et al. Common arterial trunk associated with a homeodomain mutation of NKX2.6. Hum Mol Genet 2005;14:585-93. [Crossref] [PubMed]
- Ta-Shma A, El-lahham N, Edvardson S, et al. Conotruncal malformations and absent thymus due to a deleterious NKX2-6 mutation. J Med Genet 2014;51:268-70. [Crossref] [PubMed]
- Yagi H, Furutani Y, Hamada H, et al. Role of TBX1 in human del22q11.2 syndrome. Lancet 2003;362:1366-73. [Crossref] [PubMed]
- Smemo S, Campos LC, Moskowitz IP, et al. Regulatory variation in a TBX5 enhancer leads to isolated congenital heart disease. Hum Mol Genet 2012;21:3255-63. [Crossref] [PubMed]
- Kirk EP, Sunde M, Costa MW, et al. Mutations in cardiac T-box factor gene TBX20 are associated with diverse cardiac pathologies, including defects of septation and valvulogenesis and cardiomyopathy. Am J Hum Genet 2007;81:280-91. [Crossref] [PubMed]
- Zhou YM, Dai XY, Huang RT, et al. A novel TBX20 loss-of-function mutation contributes to adult-onset dilated cardiomyopathy or congenital atrial septal defect. Mol Med Rep 2016;14:3307-14. [Crossref] [PubMed]
- Huang RT, Wang J, Xue S, et al. TBX20 loss-of-function mutation responsible for familial tetralogy of Fallot or sporadic persistent truncus arteriosus. Int J Med Sci 2017;14:323-32. [Crossref] [PubMed]
- Lu CX, Wang W, Wang Q, et al. A Novel MEF2C Loss-of-Function Mutation Associated with Congenital Double Outlet Right Ventricle. Pediatr Cardiol 2018;39:794-804. [Crossref] [PubMed]
- Abdul-Sater Z, Yehya A, Beresian J, et al. Two heterozygous mutations in NFATC1 in a patient with Tricuspid Atresia. PLoS One 2012;7:e49532 [Crossref] [PubMed]
- Ferese R, Bonetti M, Consoli F, et al. Heterozygous missense mutations in NFATC1 are associated with atrioventricular septal defect. Hum Mutat 2018;39:1428-41. [Crossref] [PubMed]
- Pizzuti A, Sarkozy A, Newton AL, et al. Mutations of ZFPM2/FOG2 gene in sporadic cases of tetralogy of Fallot. Hum Mutat 2003;22:372-7. [Crossref] [PubMed]
- De Luca A, Sarkozy A, Ferese R, et al. New mutations in ZFPM2/FOG2 gene in tetralogy of Fallot and double outlet right ventricle. Clin Genet 2011;80:184-90. [Crossref] [PubMed]
- Tan ZP, Huang C, Xu ZB, et al. Novel ZFPM2/FOG2 variants in patients with double outlet right ventricle. Clin Genet 2012;82:466-71. [Crossref] [PubMed]
- Smith KA, Joziasse IC, Chocron S, et al. Dominant-negative ALK2 allele associates with congenital heart defects. Circulation 2009;119:3062-9. [Crossref] [PubMed]
- Goldmuntz E, Bamford R, Karkera JD, et al. CFC1 mutations in patients with transposition of the great arteries and double-outlet right ventricle. Am J Hum Genet 2002;70:776-80. [Crossref] [PubMed]
- Robinson SW, Morris CD, Goldmuntz E, et al. Missense mutations in CRELD1 are associated with cardiac atrioventricular septal defects. Am J Hum Genet 2003;72:1047-52. [Crossref] [PubMed]
- Zatyka M, Priestley M, Ladusans EJ, et al. Analysis of CRELD1 as a candidate 3p25 atrioventicular septal defect locus (AVSD2). Clin Genet 2005;67:526-8. [Crossref] [PubMed]
- Maslen CL, Babcock D, Robinson SW, et al. CRELD1 mutations contribute to the occurrence of cardiac atrioventricular septal defects in Down syndrome. Am J Med Genet A 2006;140:2501-5. [Crossref] [PubMed]
- Guo Y, Shen J, Yuan L, et al. Novel CRELD1 gene mutations in patients with atrioventricular septal defect. World J Pediatr 2010;6:348-52. [Crossref] [PubMed]
- Roessler E, Ouspenskaia MV, Karkera JD, et al. Reduced NODAL signaling strength via mutation of several pathway members including FOXH1 is linked to human heart defects and holoprosencephaly. Am J Hum Genet 2008;83:18-29. [Crossref] [PubMed]
- Karkera JD, Lee JS, Roessler E, et al. Loss-of-function mutations in growth differentiation factor-1 (GDF1) are associated with congenital heart defects in humans. Am J Hum Genet 2007;81:987-94. [Crossref] [PubMed]
- Dasgupta C, Martinez AM, Zuppan CW, et al. Identification of connexin43 (alpha1) gap junction gene mutations in patients with hypoplastic left heart syndrome by denaturing gradient gel electrophoresis (DGGE). Mutat Res 2001;479:173-86. [Crossref] [PubMed]
- Wang B, Wen Q, Xie X, et al. Mutation analysis of Connexon43 gene in Chinese patients with congenital heart defects. Int J Cardiol 2010;145:487-9. [Crossref] [PubMed]
- Izumi K, Lippa AM, Wilkens A, et al. Congenital heart defects in oculodentodigital dysplasia: Report of two cases. Am J Med Genet A 2013;161A:3150-4. [Crossref] [PubMed]
- Reamon-Buettner SM, Borlak J. HEY2 mutations in malformed hearts. Hum Mutat 2006;27:118. [Crossref] [PubMed]
- Li L, Krantz ID, Deng Y, et al. Alagille syndrome is caused by mutations in human Jagged1, which encodes a ligand for Notch1. Nat Genet 1997;16:243-51. [Crossref] [PubMed]
- Oda T, Elkahloun AG, Pike BL, et al. Mutations in the human Jagged1 gene are responsible for Alagille syndrome. Nat Genet 1997;16:235-42. [Crossref] [PubMed]
- Eldadah ZA, Hamosh A, Biery NJ, et al. Familial Tetralogy of Fallot caused by mutation in the jagged1 gene. Hum Mol Genet 2001;10:163-9. [Crossref] [PubMed]
- Mohapatra B, Casey B, Li H, et al. Identification and functional characterization of NODAL rare variants in heterotaxy and isolated cardiovascular malformations. Hum Mol Genet 2009;18:861-71. [Crossref] [PubMed]
- Garg V, Muth AN, Ransom JF, et al. Mutations in NOTCH1 cause aortic valve disease. Nature 2005;437:270-4. [Crossref] [PubMed]
- Kerstjens-Frederikse WS, van de Laar IM, Vos YJ, et al. Cardiovascular malformations caused by NOTCH1 mutations do not keep left: data on 428 probands with left-sided CHD and their families. Genet Med 2016;18:914-23. [Crossref] [PubMed]
- Bleyl SB, Saijoh Y, Bax NA, et al. Dysregulation of the PDGFRA gene causes inflow tract anomalies including TAPVR: integrating evidence from human genetics and model organisms. Hum Mol Genet 2010;19:1286-301. [Crossref] [PubMed]
- Tan HL, Glen E, Töpf A, et al. Nonsynonymous variants in the SMAD6 gene predispose to congenital cardiovascular malformation. Hum Mutat 2012;33:720-7. [Crossref] [PubMed]
- Thienpont B, Zhang L, Postma AV, et al. Haploinsufficiency of TAB2 causes congenital heart defects in humans. Am J Hum Genet 2010;86:839-49. [Crossref] [PubMed]
- Reuter MS, Jobling R, Chaturvedi RR, et al. Haploinsufficiency of vascular endothelial growth factor related signaling genes is associated with tetralogy of Fallot. Genet Med 2019;21:1001-7. [Crossref] [PubMed]
- Zhao W, Wang J, Shen J, et al. Mutations in VEGFA are associated with congenital left ventricular outflow tract obstruction. Biochem Biophys Res Commun 2010;396:483-8. [Crossref] [PubMed]
- Matsson H, Eason J, Bookwalter CS, et al. Alpha-cardiac actin mutations produce atrial septal defects. Hum Mol Genet 2008;17:256-65. [Crossref] [PubMed]
- Durst R, Sauls K, Peal DS, et al. Mutations in DCHS1 cause mitral valve prolapse. Nature 2015;525:109-13. [Crossref] [PubMed]
- Ewart AK, Morris CA, Atkinson D, et al. Hemizygosity at the elastin locus in a developmental disorder, Williams syndrome. Nat Genet 1993;5:11-6. [Crossref] [PubMed]
- Metcalfe K, Rucka AK, Smoot L, et al. Elastin: mutational spectrum in supravalvular aortic stenosis. Eur J Hum Genet 2000;8:955-63. [Crossref] [PubMed]
- Micale L, Turturo MG, Fusco C, et al. Identification and characterization of seven novel mutations of elastin gene in a cohort of patients affected by supravalvular aortic stenosis. Eur J Hum Genet 2010;18:317-23. [Crossref] [PubMed]
- Li DY, Toland AE, Boak BB, et al. Elastin point mutations cause an obstructive vascular disease, supravalvular aortic stenosis. Hum Mol Genet 1997;6:1021-8. [Crossref] [PubMed]
- Ching YH, Ghosh TK, Cross SJ, et al. Mutation in myosin heavy chain 6 causes atrial septal defect. Nat Genet 2005;37:423-8. [Crossref] [PubMed]
- Posch MG, Waldmuller S, Müller M, et al. Cardiac alpha-myosin (MYH6) is the predominant sarcomeric disease gene for familial atrial septal defects. PLoS One 2011;6:e28872 [Crossref] [PubMed]
- Granados-Riveron JT, Ghosh TK, Pope M, et al. Alpha-cardiac myosin heavy chain (MYH6) mutations affecting myofibril formation are associated with congenital heart defects. Hum Mol Genet 2010;19:4007-16. [Crossref] [PubMed]
- Budde BS, Binner P, Waldmüller S, et al. Noncompaction of the ventricular myocardium is associated with a de novo mutation in the beta-myosin heavy chain gene. PLoS One 2007;2:e1362 [Crossref] [PubMed]
- Postma AV, van Engelen K, van de Meerakker J, et al. Mutations in the sarcomere gene MYH7 in Ebstein anomaly. Circ Cardiovasc Genet 2011;4:43-50. [Crossref] [PubMed]
- Zhu L, Vranckx R, Khau Van Kien P, et al. Mutations in myosin heavy chain 11 cause a syndrome associating thoracic aortic aneurysm/aortic dissection and patent ductus arteriosus. Nat Genet 2006;38:343-9. [Crossref] [PubMed]
- Olson EN. Gene regulatory networks in the evolution and development of the heart. Science 2006;313:1922-7. [Crossref] [PubMed]
- Kodo K, Nishizawa T, Furutani M, et al. Genetic analysis of essential cardiac transcription factors in 256 patients with non-syndromic congenital heart defects. Circ J 2012;76:1703-11. [Crossref] [PubMed]
- Srivastava D. Genetic regulation of cardiogenesis and congenital heart disease. Annu Rev Pathol 2006;1:199-213. [Crossref] [PubMed]
- Akçaboy MI, Cengiz FB, Inceoğlu B, et al. The effect of p.Arg25Cys alteration in NKX2-5 on conotruncal heart anomalies: mutation or polymorphism? Pediatr Cardiol 2008;29:126-9. [Crossref] [PubMed]
- Biben C, Weber R, Kesteven S, et al. Cardiac septal and valvular dysmorphogenesis in mice heterozygous for mutations in the homeobox gene Nkx2-5. Circ Res 2000;87:888-95. [Crossref] [PubMed]
- Ashraf H, Pradhan L, Chang EI, et al. A mouse model of human congenital heart disease: high incidence of diverse cardiac anomalies and ventricular noncompaction produced by heterozygous Nkx2-5 homeodomain missense mutation. Circ Cardiovasc Genet 2014;7:423-33. [Crossref] [PubMed]
- Chowdhury R, Ashraf H, Melanson M, et al. Mouse Model of Human Congenital Heart Disease: Progressive Atrioventricular Block Induced by a Heterozygous Nkx2-5 Homeodomain Missense Mutation. Circ Arrhythm Electrophysiol 2015;8:1255-64. [Crossref] [PubMed]
- Rajagopal SK, Ma Q, Obler D, et al. Spectrum of heart disease associated with murine and human GATA4 mutation. J Mol Cell Cardiol 2007;43:677-85. [Crossref] [PubMed]
- Misra C, Sachan N, McNally CR, et al. Congenital heart disease-causing Gata4 mutation displays functional deficits in vivo. PLoS Genet 2012;8:e1002690 [Crossref] [PubMed]
- LaHaye S, Majumdar U, Yasuhara J, et al. Developmental origins for semilunar valve stenosis identified in mice harboring congenital heart disease-associated GATA4 mutation. Dis Model Mech 2019;12:dmm036764 [Crossref] [PubMed]
- Wei D, Bao H, Liu XY, et al. GATA5 loss-of-function mutations underlie tetralogy of fallot. Int J Med Sci 2013;10:34-42. [Crossref] [PubMed]
- Kassab K, Hariri H, Gharibeh L, et al. GATA5 mutation homozygosity linked to a double outlet right ventricle phenotype in a Lebanese patient. Mol Genet Genomic Med 2015;4:160-71. [Crossref] [PubMed]
- Laforest B, Andelfinger G, Nemer M. Loss of Gata5 in mice leads to bicuspid aortic valve. J Clin Invest 2011;121:2876-87. [Crossref] [PubMed]
- Škorić-Milosavljević D, Tjong FVY, Barc J, et al. GATA6 mutations: Characterization of two novel patients and a comprehensive overview of the GATA6 genotypic and phenotypic spectrum. Am J Med Genet A 2019;179:1836-45. [PubMed]
- Gharibeh L, Komati H, Bossé Y, et al. GATA6 Regulates Aortic Valve Remodeling, and Its Haploinsufficiency Leads to Right-Left Type Bicuspid Aortic Valve. Circulation 2018;138:1025-38. [Crossref] [PubMed]
- Sharma A, Wasson LK, Willcox JA, et al. GATA6 mutations in hiPSCs inform mechanisms for maldevelopment of the heart, pancreas, and diaphragm. Elife 2020;9:53278. [Crossref] [PubMed]
- Basson CT, Bachinsky DR, Lin RC, et al. Mutations in human TBX5 corrected cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet 1997;15:30-5. [Crossref] [PubMed]
- Lindsay EA, Vitelli F, Su H, et al. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature 2001;410:97-101. [Crossref] [PubMed]
- Griffin HR, Töpf A, Glen E, et al. Systematic survey of variants in TBX1 in non-syndromic tetralogy of Fallot identifies a novel 57 base pair deletion that reduces transcriptional activity but finds no evidence for association with common variants. Heart 2010;96:1651-5. [Crossref] [PubMed]
- Stittrich AB, Lehman A, Bodian DL, et al. Mutations in NOTCH1 cause Adams-Oliver syndrome. Am J Hum Genet 2014;95:275-84. [Crossref] [PubMed]
- McBride KL, Riley MF, Zender GA, et al. NOTCH1 mutations in individuals with left ventricular outflow tract malformations reduce ligand-induced signaling. Hum Mol Genet 2008;17:2886-93. [Crossref] [PubMed]
- Wang B, Yan J, Mi R, et al. Forkhead box H1 (FOXH1) sequence variants in ventricular septal defect. Int J Cardiol 2010;145:83-5. [Crossref] [PubMed]
- De Luca A, Sarkozy A, Consoli F, et al. Familial transposition of the great arteries caused by multiple mutations in laterality genes. Heart 2010;96:673-7. [Crossref] [PubMed]
- Carniel E, Taylor MR, Sinagra G, et al. Alpha-myosin heavy chain: a sarcomeric gene associated with dilated and hypertrophic phenotypes of cardiomyopathy. Circulation 2005;112:54-9. [Crossref] [PubMed]
- Monserrat L, Hermida-Prieto M, Fernandez X, et al. Mutation in the alpha-cardiac actin gene associated with apical hypertrophic cardiomyopathy, left ventricular non-compaction, and septal defects. Eur Heart J 2007;28:1953-61. [Crossref] [PubMed]
- Jenkins KJ, Correa A, Feinstein JA, et al. Noninherited risk factors and congenital cardiovascular defects: current knowledge: a scientific statement from the American Heart Association Council on Cardiovascular Disease in the Young: endorsed by the American Academy of Pediatrics. Circulation 2007;115:2995-3014. [Crossref] [PubMed]
- Kuciene R, Dulskiene V. Selected environmental risk factors and congenital heart defects. Medicina (Kaunas) 2008;44:827-32. [Crossref] [PubMed]
- Lage K, Greenway SC, Rosenfeld JA, et al. Genetic and environmental risk factors in congenital heart disease functionally converge in protein networks driving heart development. Proc Natl Acad Sci U S A 2012;109:14035-40. [Crossref] [PubMed]
- Gilbert-Barness E. Teratogenic causes of malformations. Ann Clin Lab Sci 2010;40:99-114. [PubMed]
- Kalisch-Smith JI, Ved N, Sparrow DB. Environmental Risk Factors for Congenital Heart Disease. Cold Spring Harb Perspect Biol 2020;12:a037234 [Crossref] [PubMed]
- Basu M, Zhu JY, LaHaye S, et al. Epigenetic mechanisms underlying maternal diabetes-associated risk of congenital heart disease. JCI Insight 2017;2:95085. [Crossref] [PubMed]
- Basu M, Garg V. Maternal hyperglycemia and fetal cardiac development: Clinical impact and underlying mechanisms. Birth Defects Res 2018;110:1504-16. [Crossref] [PubMed]
- Chapman G, Moreau JLM. Functional genomics and gene-environment interaction highlight the complexity of congenital heart disease caused by Notch pathway variants. Hum Mol Genet 2020;29:566-79. [Crossref] [PubMed]
- Pediatric Cardiac Genomics Consortium. The Congenital Heart Disease Genetic Network Study: rationale, design, and early results. Circ Res 2013;112:698-706. [Crossref] [PubMed]
- Watkins WS, Hernandez EJ, Wesolowski S, et al. De novo and recessive forms of congenital heart disease have distinct genetic and phenotypic landscapes. Nat Commun 2019;10:4722. [Crossref] [PubMed]
- Gifford CA, Ranade SS, Samarakoon R, et al. Oligogenic inheritance of a human heart disease involving a genetic modifier. Science 2019;364:865-70. [Crossref] [PubMed]
- Brodwall K, Greve G, Leirgul E, et al. Recurrence of congenital heart defects among siblings-a nationwide study. Am J Med Genet A 2017;173:1575-85. [Crossref] [PubMed]
- Manshaei R, Merico D, Reuter MS, et al. Genes and Pathways Implicated in Tetralogy of Fallot Revealed by Ultra-Rare Variant Burden Analysis in 231 Genome Sequences. Front Genet 2020;11:957. [Crossref] [PubMed]
- Matos-Nieves A, Yasuhara J, Garg V. Another Notch in the Genetic Puzzle of Tetralogy of Fallot. Circ Res 2019;124:462-4. [Crossref] [PubMed]
- Škorić-Milosavljević D, Lahrouchi N, Bosada FM, et al. Rare variants in KDR, encoding VEGF Receptor 2, are associated with tetralogy of Fallot. Genet Med 2021; Epub ahead of print. [Crossref]
- Theis JL, Zimmermann MT, Evans JM, et al. Recessive MYH6 Mutations in Hypoplastic Left Heart With Reduced Ejection Fraction. Circ Cardiovasc Genet 2015;8:564-71. [Crossref] [PubMed]
- Tomita-Mitchell A, Stamm KD, Mahnke DK, et al. Impact of MYH6 variants in hypoplastic left heart syndrome. Physiol Genomics 2016;48:912-21. [Crossref] [PubMed]
- Theis JL, Hu JJ, Sundsbak RS, et al. Genetic Association Between Hypoplastic Left Heart Syndrome and Cardiomyopathies. Circ Genom Precis Med 2021;14:e003126 [Crossref] [PubMed]
- McKean DM, Homsy J, Wakimoto H, et al. Loss of RNA expression and allele-specific expression associated with congenital heart disease. Nat Commun 2016;7:12824. [Crossref] [PubMed]
- Jacko M, Weyn-Vanhentenryck SM, Smerdon JW, et al. Rbfox Splicing Factors Promote Neuronal Maturation and Axon Initial Segment Assembly. Neuron 2018;97:853-868.e6. [Crossref] [PubMed]
- Wei C, Qiu J, Zhou Y, et al. Repression of the Central Splicing Regulator RBFox2 Is Functionally Linked to Pressure Overload-Induced Heart Failure. Cell Rep 2015;10:1521-33. [Crossref] [PubMed]
- Verma SK, Deshmukh V, Nutter CA, et al. Rbfox2 function in RNA metabolism is impaired in hypoplastic left heart syndrome patient hearts. Sci Rep 2016;6:30896. [Crossref] [PubMed]
- Froimchuk E, Jang Y, Ge K. Histone H3 lysine 4 methyltransferase KMT2D. Gene 2017;627:337-42. [Crossref] [PubMed]
- Ng SB, Bigham AW, Buckingham KJ, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet 2010;42:790-3. [Crossref] [PubMed]
- Cuvertino S, Hartill V, Colyer A, et al. A restricted spectrum of missense KMT2D variants cause a multiple malformations disorder distinct from Kabuki syndrome. Genet Med 2020;22:867-77. [Crossref] [PubMed]
- Blue GM, Kirk EP, Giannoulatou E, et al. Targeted next-generation sequencing identifies pathogenic variants in familial congenital heart disease. J Am Coll Cardiol 2014;64:2498-506. [Crossref] [PubMed]
- Blue GM, Humphreys D, Szot J, et al. The promises and challenges of exome sequencing in familial, non-syndromic congenital heart disease. Int J Cardiol 2017;230:155-63. [Crossref] [PubMed]
- Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405-24. [Crossref] [PubMed]
- Szot JO, Cuny H, Blue GM, et al. A Screening Approach to Identify Clinically Actionable Variants Causing Congenital Heart Disease in Exome Data. Circ Genom Precis Med 2018;11:e001978 [Crossref] [PubMed]
- Sevim Bayrak C, Zhang P, Tristani-Firouzi M, et al. De novo variants in exomes of congenital heart disease patients identify risk genes and pathways. Genome Med 2020;12:9. [Crossref] [PubMed]
- Li G, Xu A, Sim S, et al. Transcriptomic Profiling Maps Anatomically Patterned Subpopulations among Single Embryonic Cardiac Cells. Dev Cell 2016;39:491-507. [Crossref] [PubMed]
- DeLaughter DM, Bick AG, Wakimoto H, et al. Single-Cell Resolution of Temporal Gene Expression during Heart Development. Dev Cell 2016;39:480-90. [Crossref] [PubMed]
- Lescroart F, Wang X, Lin X, et al. Defining the earliest step of cardiovascular lineage segregation by single-cell RNA-seq. Science 2018;359:1177-81. [Crossref] [PubMed]
- Zhang Q, Carlin D, Zhu F, et al. Unveiling Complexity and Multipotentiality of Early Heart Fields. Circ Res 2021;129:474-87. [Crossref] [PubMed]
- Lin H, McBride KL, Garg V, et al. Decoding Genetics of Congenital Heart Disease Using Patient-Derived Induced Pluripotent Stem Cells (iPSCs). Front Cell Dev Biol 2021;9:630069 [Crossref] [PubMed]
- Ge X, Ren Y, Bartulos O, et al. Modeling supravalvular aortic stenosis syndrome with human induced pluripotent stem cells. Circulation 2012;126:1695-704. [Crossref] [PubMed]
- Theodoris CV, Li M, White MP, et al. Human disease modeling reveals integrated transcriptional and epigenetic mechanisms of NOTCH1 haploinsufficiency. Cell 2015;160:1072-86. [Crossref] [PubMed]
- Ang YS, Rivas RN, Ribeiro AJS, et al. Disease Model of GATA4 Mutation Reveals Transcription Factor Cooperativity in Human Cardiogenesis. Cell 2016;167:1734-1749.e22. [Crossref] [PubMed]
- Kathiriya IS, Rao KS, Iacono G, et al. Modeling Human TBX5 Haploinsufficiency Predicts Regulatory Networks for Congenital Heart Disease. Dev Cell 2021;56:292-309.e9. [Crossref] [PubMed]
- Hrstka SC, Li X, Nelson TJ, et al. NOTCH1-Dependent Nitric Oxide Signaling Deficiency in Hypoplastic Left Heart Syndrome Revealed Through Patient-Specific Phenotypes Detected in Bioengineered Cardiogenesis. Stem Cells 2017;35:1106-19. [Crossref] [PubMed]
- Yang C, Xu Y, Yu M, et al. Induced pluripotent stem cell modelling of HLHS underlines the contribution of dysfunctional NOTCH signalling to impaired cardiogenesis. Hum Mol Genet 2017;26:3031-45. [Crossref] [PubMed]
- Miao Y, Tian L, Martin M, et al. Intrinsic Endocardial Defects Contribute to Hypoplastic Left Heart Syndrome. Cell Stem Cell 2020;27:574-589.e8. [Crossref] [PubMed]
- Paige SL, Galdos FX, Lee S, et al. Patient-Specific Induced Pluripotent Stem Cells Implicate Intrinsic Impaired Contractility in Hypoplastic Left Heart Syndrome. Circulation 2020;142:1605-8. [Crossref] [PubMed]
- Lam YY, Keung W, Chan CH, et al. Single-Cell Transcriptomics of Engineered Cardiac Tissues From Patient-Specific Induced Pluripotent Stem Cell-Derived Cardiomyocytes Reveals Abnormal Developmental Trajectory and Intrinsic Contractile Defects in Hypoplastic Right Heart Syndrome. J Am Heart Assoc 2020;9:e016528 [Crossref] [PubMed]
- Kodo K, Ong SG, Jahanbani F, et al. iPSC-derived cardiomyocytes reveal abnormal TGF-β signalling in left ventricular non-compaction cardiomyopathy. Nat Cell Biol 2016;18:1031-42. [Crossref] [PubMed]
- Kitani T, Tian L, Zhang T, et al. RNA Sequencing Analysis of Induced Pluripotent Stem Cell-Derived Cardiomyocytes From Congenital Heart Disease Patients. Circ Res 2020;126:923-5. [Crossref] [PubMed]
- Andersen JD, Jacobsen SB, Trudsø LC, et al. Whole genome and transcriptome sequencing of post-mortem cardiac tissues from sudden cardiac death victims identifies a gene regulatory variant in NEXN. Int J Legal Med 2019;133:1699-709. [Crossref] [PubMed]
- Richter F, Morton SU, Kim SW, et al. Genomic analyses implicate noncoding de novo variants in congenital heart disease. Nat Genet 2020;52:769-77. [Crossref] [PubMed]
- Landis BJ, Ware SM. The Current Landscape of Genetic Testing in Cardiovascular Malformations: Opportunities and Challenges. Front Cardiovasc Med 2016;3:22. [Crossref] [PubMed]
- Manolio TA. Bringing genome-wide association findings into clinical use. Nat Rev Genet 2013;14:549-58. [Crossref] [PubMed]
- Gaynor JW, Gerdes M, Zackai EH, et al. Apolipoprotein E genotype and neurodevelopmental sequelae of infant cardiac surgery. J Thorac Cardiovasc Surg 2003;126:1736-45. [Crossref] [PubMed]
- Gaynor JW, Kim DS, Arrington CB, et al. Validation of association of the apolipoprotein E ε2 allele with neurodevelopmental dysfunction after cardiac surgery in neonates and infants. J Thorac Cardiovasc Surg 2014;148:2560-6. [Crossref] [PubMed]
- Mital S, Chung WK, Colan SD, et al. Renin-angiotensin-aldosterone genotype influences ventricular remodeling in infants with single ventricle. Circulation 2011;123:2353-62. [Crossref] [PubMed]
- Russell MW, Chung WK, Kaltman JR, et al. Advances in the Understanding of the Genetic Determinants of Congenital Heart Disease and Their Impact on Clinical Outcomes. J Am Heart Assoc 2018;7:006906 [Crossref] [PubMed]
- Blue GM, Ip E, Walker K, et al. Genetic burden and associations with adverse neurodevelopment in neonates with congenital heart disease. Am Heart J 2018;201:33-9. [Crossref] [PubMed]
- Mercer-Rosa L, Pinto N, Yang W, et al. 22q11.2 Deletion syndrome is associated with perioperative outcome in tetralogy of Fallot. J Thorac Cardiovasc Surg 2013;146:868-73. [Crossref] [PubMed]
- Boskovski MT, Homsy J, Nathan M, et al. De Novo Damaging Variants, Clinical Phenotypes, and Post-Operative Outcomes in Congenital Heart Disease. Circ Genom Precis Med 2020;13:e002836 [Crossref] [PubMed]
- Dailey-Schwartz AL, Tadros HJ, Azamian MS, et al. Copy Number Variants of Undetermined Significance Are Not Associated with Worse Clinical Outcomes in Hypoplastic Left Heart Syndrome. J Pediatr 2018;202:206-211.e2. [Crossref] [PubMed]


