
A novel missense mutation of COL4A5 gene alter collagen IV α5 chain to cause X-linked Alport syndrome in a Chinese family
Introduction
Alport syndrome (AS) is a congenital/hereditary kidney disease with clinical features of eye, ear and kidney lesions, caused by mutations in type IV collagen genes (1). X-linked AS (XLAS), autosomal recessive AS (ARAS), and autosomal dominant AS (ADAS) are main genetic modes of inheritance AS (2,3). The most frequent form is XLAS dominant inheritance, which concerns the abnormal expression of a5 chain of type IV collagen, encoded by mutated COL4A5 gene, and account for about 85% of cases. All XLAS man will ultimately develop end-stage renal disease (ESRD) requiring dialysis or kidney transplantation by at least 60 years of age (1,4). Women with XLAS also develop proteinuria; however, only 30% develop ESRD by age 60 compared to male patients (5,6).
Thus far, more than 900 different COL4A5 mutations have been identified, such as deletions, frame shift mutations, nonsense mutations, splicing and missense mutations, which could interfere with the synthesis of the a5 chain of type IV collagen, result in loss of expression of a345 chain of type IV collagen in basement membranes (7). These mutations are spread throughout the gene without any identified mutational hot spot (8). Nighty percent of hemizygous male patients with large deletions, nonsense or small mutations progress to ESRD, whereas the probability of individuals with a missense or a splice mutation to progress to ESRD is 50–70% (9). But the detail of the mechanism for them remains unclear.
Despite so many gene variants have found to be associated with AS, there are few reports on functional verification in vitro especially in podocytes. The pathogenicity of the mutation was determined only by family validation and the prediction of protein function by software. Therefore, in this study, we report the analysis of the COL4A5 gene in a Chinese family with XLAS, and investigate the effect of a missense mutation of this family on type IV collagen.
We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tp-20-47).
Methods
Next-generation sequencing (NGS)
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics board of Shanghai Children’s Hospital (NO.2020RY049-E01) and informed consent was taken from all the patients. Genomic DNA was extracted from peripheral blood of the boy and his family members with the GentraPuregene Kit (Qiagen, Germany) with the consent of the child and his family. Targeted sequencing using NGS was conducted for genes (COL4A3/4/5) responsible for AS. The gene panel was designed using SureDesign from Agilent Technologies, including more than 200 kidney-disease genes. Gene target capture by hybridization and enrichment were performed by SureSelectXT based target enrichment system for Illumina paired-End multiplexed sequencing. The libraries were barcoded during PCR amplification after enrichment, and then were pooled and sequenced on Illumina HiSeq system. We aimed for at least 150×, and bioinformatics analysis was performed by refined pipeline on Linux system.
Sanger sequencing
In order to validate the mutation identified by NGS, PCR amplification was performed on the corresponding gene region around the mutation and Sanger sequencing was performed. The purified PCR products were sequenced using the same primers and the bidirectional PCR conditions on an ABI Genetic Analyser 3730xl (Appled Biosystems, USA) and the sequencing results were analyzed with the sequencing software (Chomas2.4.1).
Cell cultures and transfection experiments
Immortalized human podocytes AB8/13 (University of Bristol, UK) were cultured in RPMI 1640 with 10% fetal bovine serum (Gibco, USA) (10). Insulin Transferrin Selenium (Life Technologies, USA) were added to promote the proliferation of human podocytes at 33°. Cells were collected after growing to 50–60% confluency, and then subcultured to 37 °C for full differentiation in 10–14 days. The full differentiation podocytes then be used in future experiments. We purchased COL4A5-control and COL4A5-mutation plasmids, which transfected using Lipofectamine-2000 (Invitrogen), from Talen-bio Scientific Co. (Shanghai, China). Liposome-mediated transfections of podocytes were performed in four groups: none plasmid transfection (control group), empty vector plasmid pcDNA3.1(+) transfection (plasmid control group), normal COL4A5 gene-linked pcDNA3.1(+) recombinant plasmid transfected (COL4A5-normal group), pcDNA3.1(+) plasmid recombined with COL4A5 gene c.1844G>C mutant (COL4A5-mutant group). We isolated RNA and protein from the cells 36 h after transfection.
Real-time PCR
Total RNA was extracted from transfected podocytes with TRIzol reagent (Invitrogen, USA), and cDNA was generated by PrimeScript RT regent Kit (TaKaRa, Japan). The condition for reverse transcription was as follows: 15 min at 37 °C and 85 °C for 5 s. All the primers were designed as following: COL4A1_F 5'-GGGATGCTGTTGAAAGGTGAA-3', COL4A1_R 5'-GGTGGTCCGGTAAATCCTGG-3'; COL4A3_F 5'-AGCAAGGGTTGTGTCTGTAAAG-3', COL4A3_R 5'-CAGAAAATCCTGGCAATCCACT-3'; COL4A5_F 5'-CAAAAGGTGATCGTGGTTTCCC-3', COL4A5_R 5'-GTCCAGGTTGTCCATTTGGTC-3'. RT-PCR was performed using the GoTaq® qPCR Master Mix (Promeg, USA) in duplicate in 96-well plates containing 3 µL of cDNA. The cycle program was set as follows: GoTaq® hot start polymerase activation at 95 °C for 1 cycle for 2 min, denaturation at 95 °C for 15 s, annealing and extension at 60 °C for 1min for 40 cycles. The data were collected in the Light Cycler 96 (Roche, Switzerland). Relative quantitative target gene expression was performed using the 2−ΔΔCt comparative method (11) and all PCR products were replicated three times.
Western blotting
After phenylmethylsulfonyl fluoride (PMSF) pretreatment, total proteins in human podocytes were extracted by radioimmunoprecipitation assay (RIPA) buffer (Beyotime, China). Multisken MK3 (Thermo Scientific, USA) was used to measure protein concentration. Equal amounts of proteins were denatured after heating at 100 °C for 5 min and separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrohoresis (SDS-PAGE). After transferring, the polyvinylidene fluoride (PVDF) membrane (Bio-Rad, USA) was incubated with primary antibody at room temperature for 2 h and the secondary antibody for 1 h. Eventually the target bands were detected using chemiluminescence substrate (Perkin Elmer, USA). The LAS4000IR (Fujifilm, Japan) was used to capture images. The expression of proteins was analysed with the ImageReader LAS-4000 software. Rabbit anti-human COL4A5 (Santa Cruz Biotechnology, Cat. # sc-11360, USA) has been used with a 1:500 dilution; COL4A3 (Abcam, Cat. # ab73981, UK) and goat anti-human COL4A1 (Novus, Cat. # NBP1-26549, USA) have been used with a 1:1,000 dilution; beta-actin, mouse anti-β-Actin monoclonal antibody (Santa Cruz Biotechnology, Cat. # sc-47778, RRID: AB_626632, USA) have been used with a 1:20000 dilution; goat anti-rabbit IgG-HRP (Jackson, Cat. # 111-035-003, RRID: AB_2313567, UK) and goat anti-mouse IgG-HRP linked antibody (Yeasen-bio Technology, Cat. # 33201ES60, China) have been used with a 1:10,000 dilution. Western Blot strips were quantitatively analyzed by using Image J software to obtain the gray value. The final results were expressed by the ratio of the average gray value of COL4A1, COL4A3 and COL4A5 to the average gray value of beta-actin.
Immunofluorescence analysis
Culture dishes were added with 4% paraformaldehyde for 30min to fix the podocytes, and then cells were blocked with 2% BSA at room temperature for 30 min. After three times of PBS washing, podocytes were incubated with rabbit anti-human COL4A3 (Sigma-Aldrich, Cat. # SAB4500376, RRID: AB_10742910, USA) and COL4A5 antibody (Sigma-Aldrich, Cat. # SAB4500381, RRID: AB_10743809, USA, all diluted at 1:500, 37 °C for 1 h) and goat anti-human COL4A1 antibody (Novus, Cat. # NBP1-26549, USA, diluted at 1:500, 37 °C for 1 h). Goat-anti-Rabbit-Alexa Flour 488 (Abcam, Cat. # ab150077, RRID: AB_2630356, UK) and Goat-anti-Mouse Alexa Flour 561 (Abcam, Cat. # ab150113, UK) was added (diluted at 1:1,000, room temperature for 1 h) and washed respectively. Fluoroshield Mounting Medium with DAPI (Thermo Fisher Scientific Cat# D3571, RRID: AB_2307445, USA) was used to stain nuclei (diluted at 1:10,000, room temperature for 10 min). Cells were visualized by confocal microscopy (Leica, Germany). Image J software was used to analyze the mean fluorescence intensity of target proteins (COL4A1, COL4A3 and COL4A5) in cells.
Statistical analysis
SPSS 17.0 version was used for statistical analysis, and the data were expressed as mean ± standard deviation (SD). One way analysis of variance (ANOVA) were used for intergroup comparisons, followed by a Least-Significant Difference test. P<0.05 was considered statistically significant.
Results
Case report
The patient was a 10-year-old Chinese boy who was diagnosed with gross hematuria and proteinuria after a respiratory tract infection 3 year ago. Urinalysis revealed microscopic hematuria (full view/high-power field of urine blood cells), urine protein of + to 2+, and urine protein level of 0.34 g/24 h. Blood tests revealed blood urea nitrogen of 5.8 mmol/L and creatinine of 41 µmol/L, indicating preserved renal function. His blood pressure was normal and no ocular disorder or hearing loss was found. His mother tested microscopic hematuria (45–55/high-power field of urine blood cells) after his AS onset without renal failure, his father and other family members were tested normal. Renal biopsy showed a diagnosis of minor glomerular abnormalities, and immunofluorescence showed normal expression of type IV collagen a1 and a2 chains and no expression of a5 chain in the glomerular basement membrane (GBM) (Figure 1). However, he didn’t receive an electron microscope.
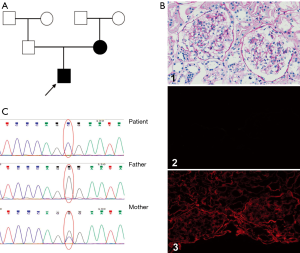
NGS identified potentially pathogenic AS mutation
With the consent of the patient and his family (the trial was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics board of Shanghai Children’s Hospital (No. 2020RY049-E01) and informed consent was taken from all the patients), we performed genetic analysis and found a novel missense mutation c.1844G>C in exon 25 of COL4A5 (NM_000495) (Figure 1B). No mutation in the COL4A3/4 genes of this patient was found. The patient’s mother also had the same mutation in exon 25, while his father was normal. The frequency of this site in the population is extremely low, and there are no relevant reports on pathogenicity of this site, while its clinical significance was unknown. The protein function predicted by Sorts intolerant from tolerant (SIFT) and polymorphism phenotyping (PolyPhen) was harmful.
The c.1844G>C variant in COL4A5 affects its mRNA expression in podocytes
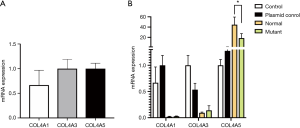
In this experiment, we found that the expression of COL4A1/3/5 mRNA could be detected in normal human podocytes, and the expressions of a3 chain and a5 chain are equivalent, higher than a1chain.
Four groups (control group, plasmid control group, COL4A5-normal and COL4A5-mutant groups) were transiently transfected into podocytes, and COL4A5 mRNA level was analyzed by real-time PCR. We found the COL4A5 mRNA expression of COL4A5-normal and COL4A5-mutant group were significantly higher than that of the plasmid control group (P<0.01), and compared with COL4A5-normal, the mRNA level of COL4A5-mutant group was higher (P<0.05). However, pairwise-comparison showed that there was no significant difference of the COL4A1 and COL4A3 mRNA expression between the four groups (Figure 2).
The c.1844G>C variant in COL4A5 reduced a5 chain of type IV collagen expression in podocytes
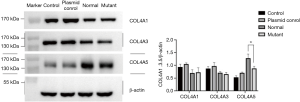
After 36 h podocyte transfection, western blot was used to detect COL4A5 protein expression level in all groups. Consistent with the results of real-time PCR, we also detected the expression of COL4A5-mutant group was significantly reduced than that of COL4A5-normal and plasmid control groups (P<0.01), and there was no significant difference of a1/3 chain of type IV collagen expression between all groups (Figure 3).
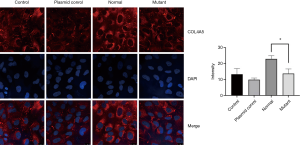
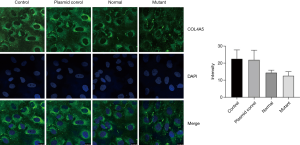
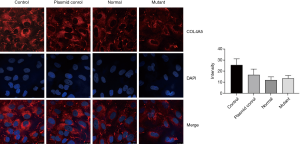
We further use immunofluorescence to detect the COL4A5 expression in podocytes. It is obvious that after transfection the fluorescence intensity of COL4A5-normal group was significantly increased than the control and plasmid control groups, but the expression of COL4A5 significantly reduced than the normal group after transfection of the mutant plasmid (Figure 4). Again, there was no significant difference in COL4A3 and COL4A1 protein immunofluorescence staining between the normal and mutant groups (Figures 5,6).
Discussion
In this study, we described a novel COL4A5 variant in an AS family in which both males and females are affected. The condition of the proband boy was more serious than that of his mother, such as more severe and progressive hematuria and proteinuria, and renal biopsy showed no expression of a5 (IV) in GBM. Furthermore, genetic testing revealed a hemizygote missense mutation c.1844C>A in exon 25 of COL4A5 gene both in the boy and his mother, all above suggesting the diagnoses of classic X-linked pattern of inheritance AS.
COL4A5 gene contains 51 exons and 50 introns, encoding 1,685 amino acids of type IV collagen a5 chain in GBM, which is one of the most important parts of GBM (12,13). Previous studies have shown that COL4A5 protein function changes can be induced after COL4A5 gene mutation in XLAS patients, which further preventing to detect a5 (IV) in GBM of most patients (14). Thus far, more than 900 different variants spreading throughout the COL4A5 gene have been identified without any identified mutational hot spot (7). Whether these variants can influence the function of COL4A5 protein is usually predicted by some software, such as SIFT and Kashtan et al. (7), but rarely confirmed by in vitro experiments. In this research, COL4A5 mRNA expression level in the mutant group was significantly lower than that in the normal group, indicating that this variant was probably affected the gene transcription. And further western blot and immunofluorescence results showed that COL4A5 c.1844G>C variant significantly down-regulated COL4A5 protein expression in podocytes after transfection. a5 (IV) is only synthesized by podocytes (15), the variant may be the main cause of a5 (IV) deletion of GBM, which is a pathogenic mutation of the patient.
Missense mutation was the most mutation types of COL4A5 gene, accounted for 43.6% of all mutations in the AS COL4A5 Variant Database (16) (http://www.arup.utah.edu/database/ALPORT/ALPORT_welcome.php). Half of patients with missense mutations were reported to enter ESRD by age 30, and have a 50% renal survival rate of 32 years, and in some family with missense mutations, both hearing loss and ESRD occurred after age of 50 (5,17). This is different from patients with nonsense, deletion, or insertion mutations, which change the reading frame of the gene. 90% male patients with these mutations tend to progress to ESRD before age 30, and the kidney survival rate before age 20 is only 50% (5). Therefore, further research is needed to determine whether missense mutation of COL4A5 gene will cause changes in protein function and thus cause AS. However, our experiment can verify the effectiveness of COL4A5 mutation through in vitro experiments, so as to determine whether this is a pathogenic gene.
Meanwhile, western blot, immunofluorescence and real-time PCR were also used to detect COL4A3 and COL4A1 protein and mRNA levels in podocytes in each group, but no significant differences were found. Collagen IV has 6 α-chains, α1 to α6 chains, encoded by the genes COL4A1-COL4A6: COL4A1 and COL4A2 on chromosome 13; COL4A3 and COL4A4 on chromosome 2; and COL4A5 and COL4A6 on chromosome X respectively (18). Each collagen protein is composed of N-terminal 7S domain, intermediate Gly-x-y repeats and non-collagenous (NC1) domain at the C-terminus (18). The six collagen IV a-chains formed α1α1α2, a3a4a5 and α5α5α6 network comprising helical heterotrimers, which primarily produced by podocytes and defines the mature GBM (8,15,18-20). Mutations in any of the three type IV collagen chains of a3a4a5 network often inhibit the expression of the other two chains and the formation of trimers in the GBM (21). COL4A3 gene knockout mice don’t express neither a4 (IV) nor a5 (IV) chains in their glomeruli (22). However, Korstanje et al. found that α3, α4, and α5 (IV) chains can still be detectable in GBM in a COL4A4 mutant AS mouse model (23). In our research, the patient’s renal biopsy showed no expression of a3 (IV) and a5 (IV) in GBM, therefore, the down-regulated expression of a5 (IV) chain may prevent the inability of the collagen a3a4a5 (IV) trimers network forming and lead to abnormal human GBM structure, so as to destroy the glomerular filtration barrier, leading to a large number of red blood cells and albumin leakage, progressive loss of kidney function, eventually to end-stage renal disease. But the specific mechanism still needs further study.
Conclusions
In conclusion, this study reports a novel missense variant, c.1844G>C, in exon 25 of COL4A5 gene in a family with XLAS. And it may down-regulate the expression level of a5 (IV), thus leading to the deletion of type IV collagen chains of a3a4a5 network in GBM in this Chinese boy.
Acknowledgments
Funding: This research was funded by the Training Program Foundation for the Talents by the Shanghai Municipal Commission of Health and Family Planning (grant number 2018YQ21), the National Natural Science Foundation of China (grant number 81500540), and Youth Program Foundation by the Shanghai Municipal Commission of Health and Family Planning (grant number 20164Y0192).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tp-20-47
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tp-20-47
Peer Review File: Available at http://dx.doi.org/10.21037/tp-20-47
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tp-20-47). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by institutional ethics board of Shanghai Children’s Hospital (No. 2020RY049-E01) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Gillion V, Dahan K, Cosyns JP, et al. Genotype and Outcome After Kidney Transplantation in Alport Syndrome. Kidney Int Rep 2018;3:652-60. [Crossref] [PubMed]
- Savige J. Alport syndrome: its effects on the glomerular filtration barrier and implications for future treatment. J Physiol 2014;592:4013-23. [Crossref] [PubMed]
- Kuebler B, Aran B, Miquel-Serra L, et al. Generation of integration-free induced pluripotent stem cell lines derived from two patients with X-linked Alport syndrome (XLAS). Stem Cell Res 2017;25:291-5. [Crossref] [PubMed]
- Voskarides K, Papagregoriou G, Hadjipanagi D, et al. COL4A5 and LAMA5 variants co-inherited in familial hematuria: digenic inheritance or genetic modifier effect? BMC Nephrol 2018;19:114. [Crossref] [PubMed]
- Jais JP, Knebelmann B, Giatras I, et al. X-linked Alport syndrome: natural history in 195 families and genotype- phenotype correlations in males. J Am Soc Nephrol 2000;11:649-57. [PubMed]
- Bekheirnia MR, Reed B, Gregory MC, et al. Genotype-phenotype correlation in X-linked Alport syndrome. J Am Soc Nephrol 2010;21:876-83. [Crossref] [PubMed]
- Kashtan CE, Ding J, Garosi G, et al. Alport syndrome: a unified classification of genetic disorders of collagen IV alpha345: a position paper of the Alport Syndrome Classification Working Group. Kidney Int 2018;93:1045-51. [Crossref] [PubMed]
- Liu JH, Wei XX, Li A, et al. Novel mutations in COL4A3, COL4A4, and COL4A5 in Chinese patients with Alport Syndrome. PLoS One 2017;12:e0177685. [Crossref] [PubMed]
- Gotenstein JR, Koo CC, Ho TW, et al. Genetic Suppression of Basement Membrane Defects in Caenorhabditis elegans by Gain of Function in Extracellular Matrix and Cell-Matrix Attachment Genes. Genetics 2018;208:1499-512. [Crossref] [PubMed]
- Saleem MA, O'Hare MJ, Reiser J, et al. A conditionally immortalized human podocyte cell line demonstrating nephrin and podocin expression. J Am Soc Nephrol 2002;13:630-8. [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001;25:402-8. [Crossref] [PubMed]
- Zhou J, Leinonen A, Tryggvason K. Structure of the human type IV collagen COL4A5 gene. J Biol Chem 1994;269:6608-14. [PubMed]
- Barua M, John R, Stella L, et al. X-Linked Glomerulopathy Due to COL4A5 Founder Variant. Am J Kidney Dis 2018;71:441-5. [Crossref] [PubMed]
- Savige J, Gregory M, Gross O, et al. Expert guidelines for the management of Alport syndrome and thin basement membrane nephropathy. J Am Soc Nephrol 2013;24:364-75. [Crossref] [PubMed]
- Cosgrove D, Liu S. Collagen IV diseases: A focus on the glomerular basement membrane in Alport syndrome. Matrix Biol 2017;57-58:45-54. [Crossref] [PubMed]
- International Alport Mutation C, Savige J, Ars E, et al. DNA variant databases improve test accuracy and phenotype prediction in Alport syndrome. Pediatr Nephrol 2014;29:971-7. [Crossref] [PubMed]
- Barker DF, Pruchno CJ, Jiang X, et al. A mutation causing Alport syndrome with tardive hearing loss is common in the western United States. Am J Hum Genet 1996;58:1157-65. [PubMed]
- Chew C, Lennon R. Basement Membrane Defects in Genetic Kidney Diseases. Front Pediatr 2018;6:11. [Crossref] [PubMed]
- Borza DB, Bondar O, Ninomiya Y, et al. The NC1 domain of collagen IV encodes a novel network composed of the alpha 1, alpha 2, alpha 5, and alpha 6 chains in smooth muscle basement membranes. J Biol Chem 2001;276:28532-40. [Crossref] [PubMed]
- Abrahamson DR, Hudson BG, Stroganova L, et al. Cellular origins of type IV collagen networks in developing glomeruli. J Am Soc Nephrol 2009;20:1471-9. [Crossref] [PubMed]
- Massella L, Gangemi C, Giannakakis K, et al. Prognostic value of glomerular collagen IV immunofluorescence studies in male patients with X-linked Alport syndrome. Clin J Am Soc Nephrol 2013;8:749-55. [Crossref] [PubMed]
- Cosgrove D, Meehan DT, Grunkemeyer JA, et al. Collagen COL4A3 knockout: a mouse model for autosomal Alport syndrome. Genes Dev 1996;10:2981-92. [Crossref] [PubMed]
- Korstanje R, Caputo CR, Doty RA, et al. A mouse Col4a4 mutation causing Alport glomerulosclerosis with abnormal collagen alpha3alpha4alpha5(IV) trimers. Kidney Int 2014;85:1461-8. [Crossref] [PubMed]


