
Tumor dormancy is closely related to prognosis prediction and tumor immunity in neuroblastoma
Highlight box
Key findings
• A 6-gene genetic signature for neuroblastoma-based on dormancy could predict the prognosis of patients and response to immunotherapy.
What is known and what is new?
• Most high-risk neuroblastomas present with widespread metastatic disease at diagnosis and either do not respond to conventional therapies initially or ultimately relapse after treatment. In this process, tumor cell dormancy plays an important role.
• The present study is the first to incorporate a dormancy-associated signature for predicting NB survival and the response to immunotherapy. The nomogram would have certain guiding significance in decision-making for clinical treatment.
What is the implication, and what should change now?
• In this study, a dormancy-associated gene signature, including CDKN2A, BHLHB3, CDKN2B, MAPK14, CDKN1B, and BMP7, was established. The gene signature showed a strong capacity to predict NB patient prognosis and had certain guiding significance in decision-making for clinical treatment.
Introduction
Neuroblastoma (NB), which is the most frequent and fatal solid tumor in early childhood, is characterized by heterogeneous behavior from spontaneous regression to relentless progression (1). Despite early diagnosis and new approaches to treatment, NB relapse remains one of the biggest clinical challenges in the field and adversely affects survival (2). In a significant proportion of children with NB, distant metastases emerge after years or even decades of latency. Among individuals with high-risk NB, relapse occurs in over 50% of cases, and the 5-year survival rate is less than 40% (3). A number of biomarkers have provided guidance to physicians regarding treatment and prognosis of NB, including International Neuroblastoma Staging System (INSS) stage, MYCN (N-myc, myelocytomatosis oncogene) status, age at diagnosis, histologic category, grade of differentiation, mitosis-karyorrhexis index (MKI), and DNA ploidy. However, none of these biomarkers can accurately predict recurrence (4,5).
Despite progress in intensive multimodal treatment, most high-risk NBs present with widespread metastatic disease at diagnosis and either do not respond to conventional therapies initially or ultimately relapse after treatment, with long-term survival less than 50% (3). Immunotherapy can greatly improve the survival rate of cancer patients while reducing acute or chronic toxicities, and it is emerging as a viable alternative or adjuvant to current treatments for high-risk NB. Immunotherapy has successfully changed high-risk NB from a uniformly fatal disease to a potentially curable one in more than half of patients (6). However, challenges remain in optimizing and expanding immunotherapeutic targets, and a better understanding of the opportunities and response to immunotherapy is critical in shaping new NB treatment perspectives.
Although relapse and metastasis are the leading causes of NB-related mortality, the underlying mechanisms remain poorly understood. Dormant disseminated tumor cells (DTCs) have been reported in NB (7). DTCs are a subpopulation of circulating tumor cells (CTC) and a portion of disseminated tumor cells that engaged in metastatic niches (8). Generally, the ability to maintain viability over long periods in an equilibrium between proliferation and death is referred to as a state of tumor dormancy. There are three hypothesized models of dormancy: (I) cellular dormancy, the most accepted model, in which cells are characterized as G0/G1 cell cycle arrest achieved by activation of quiescence programs; (II) angiogenic dormancy, in which cells remain separated in surroundings with a low concentration of oxygen and nutrients; and (III) immunosurveillance dormancy, in which the immune system prevents tumoral re-growth (9). p38 MAPKhigh/ERKlow phenotype is now generally labelled as a marker of dormant cells, furthermore, some proliferation marker and the cyclin-dependent kinase (CDK) inhibitor are often used to identify dormant tumor cells (7). After the activation of dormant tumor cells, metastatic recurrences occurred months, years, or even decades after the initial radical treatment. Dormant NB cells are responsible for tumor-promoting activity, including tumor cell growth, migration, metastasis, and drug resistance as well as suppressive activity in the immune system. Nonetheless, a comprehensive investigation of the relationship between dormancy and NB prognosis and its potential association with tumor immunity has yet to be conducted.
Nomogram prognostic models that predict NB prognosis in children may facilitate better treatment decisions and assessments. Our present study showed that a 6-gene genetic signature for NB-based on dormancy could predict prognosis of neuroblastoma patients and response to immunotherapy. We present the following article in accordance with the TRIPOD reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-23-119/rc).
Methods
Obtaining of expression and clinical data
Clinical data and gene expression datasets for NB were retrieved from the Gene Expression Omnibus (GEO) under the accession number GSE49710 (Zhang et al., n=498) (10) and the ArrayExpress database under the accession number E-MTAB-8248 (Roderwieser et al., n=223) (11). The GSE49710 dataset was used to construct the prognostic model, which was then validated using the E-MTAB-8248 dataset.
Consensus clustering
Dormancy-associated genes were obtained from the existing literature. Expression data of dormancy-associated genes, normalized by the median value before clustering, were extracted from the GSE49710 dataset. The “ConsensusClusterPlus” package was used to cluster NB samples (12).
Differentially expressed gene (DEG) identification
The NB samples were divided into different groups through clustering. We used the documentation provided by the manufacturer to annotate the probe matching genetic symbols. When a single gene matched more than 1 probe, the highest expression value was recommended to represent the gene expression level. We utilized the “limma” package (13) to identify DEGs between groups (13). The DEG cutoffs were set as |log2 [fold-change ([FC)]| > 1 and adjusted P value <0.01.
Functional enrichment analysis of DEGs
Functional analyses of DEGs, consisting of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses, were performed using the “ClusterProfiler” package (14). P<0.05 was considered statistically significant.
Assessment of tumor immunity
The ESTIMATE method (Estimation of Stromal and Immunological cells in Malignant Tumor tissues utilizing Expression data) was used to determine the stromal, immune, and ESTIMATE scores (15). The microenvironment cell populations-counter (MCP-Count) method was used to rate eight immune cells (16).
Establishment of predictive dormancy signature
The identification of dormancy genes associated with overall survival was performed using Cox proportional hazards regression analysis. Genes with P<0.01 were considered statistically significant in the univariate Cox regression analysis and included in consecutive analysis. The identified genes were further optimized using the multivariate Cox regression analysis through the minimum Akaike information criterion (AIC) value. A linear combination of the multivariate Cox regression model’s coefficients and its mRNA expression level was employed to create. According to the median risk score, a 6-gene signature was developed and used to divide the NB patients into a high- or low-risk group, which was evaluated using Kaplan-Meier analysis, the area under the curve (AUC) of the receiver operating characteristic (ROC) curve, and Harrell’s concordance index (C-index). Validation of the signature was conducted using the E-MTAB-8248 dataset, and the risk score was calculated using the same formula as that for the GSE49710 dataset. A threshold value calculated from the GSE49710 dataset was used to divide patients into low-risk and high-risk groups, and the 2 groups were then compared to evaluate the prognostic model.
The identification of NB independent prognostic characteristics
On the basis of the predictive signature and known clinicopathological risk factors such as gender, age, INSS stage, and MYCN status, a univariate Cox regression analysis was performed. The significance level was set at P<0.05.
Construction of predictive nomogram
Following collinearity testing, all independent prognostic characteristics were considered. After testing the proportional hazards assumption through the Schoenfeld residuals in the “survminer” package, the characteristics were included in the construction of a prognostic nomogram to predict 1-, 3-, and 5-year overall survival of NB. In addition to a calibration plot, the performance of the nomogram was assessed using the AUC of the ROC curve and Harrell’s C-index. On the basis of nomogram points, we divided the patients into 3 groups and analyzed their survival curves using Kaplan-Meier analysis.
Ethics
This study was approved by the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital (No. E20210027) and it was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Written informed consent was obtained from the parents or legal guardians of each patient.
Patients and tissue specimens
Tissue samples from primary peripheral NB tumors were tested and verified by two independent pathologists. Collection and manipulation of human samples from 24 patients hospitalized from November, 2015 to October, 2020.
Western blotting and antibodies
The tissues were put into centrifuge tube to grind into tissue homogenate lysed with RIPA lysis buffer supplemented with 1 mM Na3VO4, 1 mM NaF and phosphatase inhibitor cocktail (Hoffman-la Roche Ltd, Basel, Switzerland). The protein was denatured in a 95 ℃ water bath and centrifuged at 12,000 rpm at 4 ℃ for 10 min, respectively. Then the upper clear protein lysates were collected. The steps for WB were based on previously described methods. The following antibodies were used: anti-CDKN2A (1:1,000) and anti-GAPDH (1:1,000) from Bioss (Beijing, China).
Reverse transcription-polymerase chain reaction (RT-PCR)
Primary tumors were lysed with TRIzol reagent (Ambion, TX, USA) for total ribonucleic acid (RNA) purification. SuperScript II Reverse Transcriptase was utilised for reverse transcription (Invitrogen). Primer sequences are listed below: CDKN2A: F-primer: GCTGCTCACCTCTGGTGCCAAA, R-primer: ACCTGCGCACCATGTTCTCG; BMP7: F-primer: ACGAGGTGCACTCGAGCTT, R-primer: GAAGCGTTCCCGGATGTAGT. The amplification reaction was carried out using primers designed in accordance with the manufacturer’s instructions (Takara, Kusatsu, Japan).
Statistical analysis
RStudio (version 1.1.463) was used to conduct the statistical analysis. Kaplan-Meier analysis was carried out to compare subgroups of patients. We conducted univariate Cox and multivariate Cox regression analyses to evaluate overall survival-related characteristics. Immune cells among subgroups were compared with the Mann-Whitney test. Statistical significance was defined as P<0.05, unless otherwise specified.
Results
NB classification based on dormancy-related genes
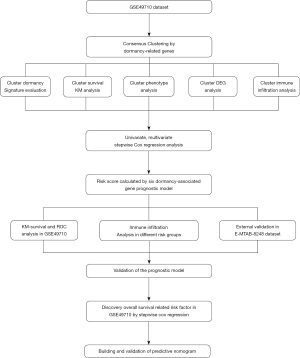
The flow chart of the research design is shown in Figure 1. A total of 20 dormancy-associated genes were extracted from the existing literature, 19 of which were expressed in the GSE49710 dataset. We conducted consensus clustering analysis using dormancy gene expression in the “ConsensusClusterPlus” package to further explore the internal relationship between dormancy and NB. By applying the clustering variable (k) from 2 to 8, we observed that k=2 showed outstanding performance, with low intergroup correlations and high intragroup correlations. Based on the clustering results, the 498 NB samples in the GSE49710 dataset constituted 2 clusters, 1 containing 256 samples and the other 242 samples (Figure 2A). The 2 clusters showed considerable separation in t-distributed stochastic neighbor embedding (t-SNE) analysis (Figure 2B). Box plots of the dormancy signature score distribution calculated by 3 different methods [single-sample Gene Set Enrichment Analysis (ssGSEA), principal component analysis (PCA), and the z-score algorithm] implied that cluster 2 had a higher dormancy score (Figure 2C-2E). Kaplan-Meier survival analysis showed that samples in cluster 2 exhibited significantly worse prognosis (P<0.0001, Figure 2F). To further explore the correlation between clusters and dormancy-related phenotypes, dormancy-associated markers which had previously been reported for their association with tumor dormancy, including DNA replication, cell cycle transition and phases, mitotic spindle formation, and glucose metabolism, were scored using the PCA algorithm, with the results exhibiting statistically significant differences (P<0.05, Figure 2G-2L). However, apoptosis showed no significant difference between 2 clusters (P>0.05, Figure 2M).
Exploration of cluster character
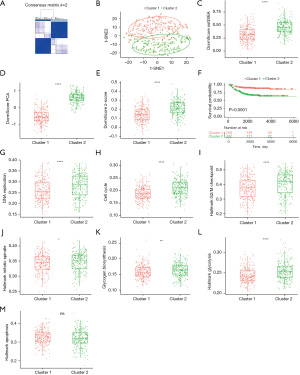
Stromal and immune cell infiltration was evaluated using the ESTIMATE algorithm to investigate differences in tumor immunity between clusters. In comparison with cluster 2, cluster 1 was found to have a significantly higher immune score, stromal score, and ESTIMATE score, suggesting a greater infiltration of immune cell and stromal cell in cluster 1 (Figure 3A-3C). To further elucidate the tumor-infiltrating immune cells, MCP-Count was utilized, with the results showing that CD8+ T cells and dendritic cells were more highly infiltrated in cluster 1 (Figure 3D), implying that there may be an increased immune cell infiltration and activate the immune response in the tumor microenvironment if the dormancy signature is higher. Further exploration of the interleukin-related signatures was made in the 2 clusters, and cluster 1 showed significantly higher levels (Figure 3E,3F).
To determine the pathway differences between clusters, we analyzed the DEGs between clusters with the following criteria: |logFC|>1 and adjusted P<0.01. A total of 31 upregulated and 37 downregulated genes were identified between the 2 clusters (Figure S1A). Interestingly, the well-known CHD5 tumor suppressor gene in NB was included in the downregulated genes, which was consistent with a previous study (17). The biological function of DEGs was explored through GO enrichment analysis, which showed synapse organization was an area of significant enrichment for the DEGs (Figure S1B). Furthermore, GSEA revealed cytokine-related signaling pathways involved in the dormancy process (Figure S1C).
Identification of survival-related dormancy signature genes and establishment of dormancy 6-gene prognostic signature
The 498 samples in the GSE49710 dataset containing sufficient overall survival information were included in the subsequent Cox regression analysis. The clinical information of these patients is presented in Table S1. Based on univariate Cox regression analysis, 7 dormancy-related genes were significantly associated with overall survival (P<0.01). BMP7 and CDKN2A manifested a hazard ratio (HR) >1 and may have contributed to tumor promotion, while other genes evidenced by HR <1 may have served as tumor protective factors in NB (Figure 4A). The results of Kaplan-Meier survival analysis showed all 7 dormancy-associated genes had significantly different survival rates (Table S2, log-rank P<0.01). A dormancy-related prognostic signature comprising 6 genes, including CDKN2A, BHLHB3, CDKN2B, MAPK14, CDKN1B, and BMP7, was developed through multivariate Cox regression analysis (Figure 4B). The following equation was used to calculate the risk score:
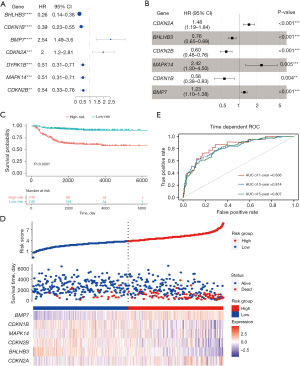
The threshold was established as the median risk score (3.33). The patients in the GSE49710 dataset could be divided into 2 distinct groups, and patients with low-risk scores showed a significantly more favorable overall survival in the Kaplan-Meier survival curves (P<0.0001, Figure 4C,4D). Time-dependent ROC and C-index were then used to evaluate the 6-gene model. The model achieved AUCs of 0.836, 0.814, and 0.807 for predicting overall survival at 1 year, 3 years, and 5 years, respectively (Figure 4E). The C-index of the 6-gene model was 0.773 (95% CI: 0.730−0.816), indicating that the 6-gene signature performed well in predicting NB outcome.
External validation of prognostic performance of the 6-gene signature
The performance of the 6-gene signature was validated using the E-MTAB-8248 dataset. The risk score was calculated following the same equation. Patients were divided into high-risk and low-risk groups based on the median risk score calculated from the GSE49710 dataset. The t-SNE plot showed a considerable separation for the 2 risk groups (Figure 5A). Moreover, there was a substantial difference in overall survival, as seen by the Kaplan-Meier survival curve. Compared with the low-risk group, patients in the high-risk group had poorer outcomes (P<0.0001, Figure 5B,5C). Time-dependent ROC and C-index analyses were then used to assess the model’s prediction performance. In the E-MTAB-8248 dataset, the AUCs for 1-, 3-, and 5-year overall survival predictions for the risk scores were 0.711, 0.815, and 0.812, respectively (Figure 5D). External validation provided further confirmation of the predictive ability of the 6-gene signature.

Clinical-pathology and tumor immunity of the 6-gene signature
Associations between the 6-gene signature and the clinical characteristics of NB, including INSS stage, MYCN status, tumor progression, and lactate dehydrogenase (LDH) and neurotrophic tyrosine receptor kinase (NTRK) expression, were analyzed in the GSE49710 dataset. In terms of the INSS stage, patients with stages 1 and 2 had lower risk scores than those with stages 3 and 4. From stage 1 to 4, the risk score increased gradually (Figure 6A). MYCN amplification and progression were more likely to occur in patients with high-risk scores (Figure 6B,6C). Furthermore, patients with high-risk scores tended to have high expression of LDH and low expression of NTRK, which correlated with a poor prognosis of NB (Figure 6D-6F). Based on the ESTIMATE algorithm, we analyzed stromal and immune cell infiltration for differences in tumor immunity. The low-risk group showed significantly higher immune scores, stromal scores, and ESTIMATE scores compared with the high-risk group, indicating increased immune-cell and stromal infiltration (Figure 6G-6I). The MCP-Count algorithm was used to quantify tumor-infiltrating immune cells, and in the low-risk group, antitumor immune cells, including cytotoxic lymphocytes and natural killer cells, were more highly infiltrated (Figure 6J).

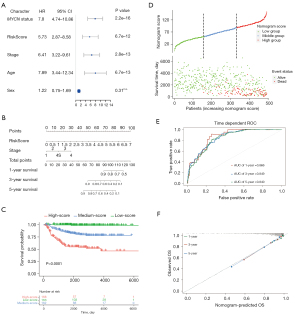
Evaluation of prognostic factors in NB and nomogram construction
Univariate and multivariate Cox regression analyses were performed to identify independent prognostic factors in a cohort of 493 patients from the GSE49710 dataset with complete clinical information, including age, sex, MYCN status, and INSS stage. Univariate Cox analysis showed that risk score (P=6.7e-12), age (P=6.7e-13), INSS stage (P=2.8e-13), and MYCN status (P=2.2e-16) were significantly correlated with overall survival (Figure 7A). After testing the proportional hazards assumption, the risk score and INSS stage were included in the construction of a nomogram to predict outcomes for 1 year, 3 years, and 5 years (Figure 7B). Patients were classified into 3 groups based on the points of the nomogram. A Kaplan-Meier plot effectively distinguished samples of different risk groups, with the low-risk group having a significantly better outcome (P<0.0001, Figure 7C,7D). The AUCs of the 1-, 3-, and 5-year overall survival predictions for the nomogram model were 0.866, 0.849, and 0.840, respectively (Figure 7E). Similar conclusions were obtained from analysis of the external dataset (E-MTAB-8248) (Figure S2A,S2B). The C-index of the nomogram model was 0.859 (95% CI: 0.832−0.886). Additionally, the 1-, 3-, and 5-year calibration plots demonstrated that the nomogram could accurately predict the outcome of NB (Figure 7F).
To further clarify the role of CDKN2A and BMP7, 24 samples of NB tissues were gathered from patients who had received curative surgery between 2015 and 2020 at Tianjin Medical University Cancer Institute and Hospital. This cohort of NB patients was enrolled and divided into early (<2 years) and late (≥2 years) recurrence groups. We first analyzed the mRNA expression of CDKN2A and BMP7 in human NB samples, with the results showing conspicuously higher CDKN2A and BMP7 expression in tumor tissues of the late recurrence group (Figure S3A). The CDKN2A protein level was also remarkably upregulated in NB tissues of the late recurrence group (Figure S3B). Although not all of the molecules in the 6-gene signature and nomogram model have been confirmed, we will further optimize this model in future clinical work in order to improve the prognosis of patients to the greatest extent.
Discussion
NB causes significant tumor-associated mortality worldwide. Due to its heterogeneity, not all patients respond to targeted therapies, and subtherapeutic immunotherapy and drug resistance are common. Although there are methods now available to predict NB prognosis such as INSS, histologic category, and DNA ploidy, there is still a lack of effectively accurate method to prevent or predict recurrence. Therefore, the development of a more precise and satisfactorily predictive model for treatment evaluation as well as overall survival of NB is urgently needed.
Tumor dormancy is a reversible state of cellular quiescence with a duration ranging from months to prolonged periods of time when tumors remain undetectable before becoming an overtly progressive disease. Tumor dormancy has been found to be implicated in therapy resistance, relapse, and metastasis in multiple tumors and is one of the deadliest features of cancer (9). In a study of NB, researchers have verified that dormancy is responsible for therapeutic resistance to most current antitumor agents and is associated with metastasis, recurrence, and increased morbidity (18). Thus, it is of great significance to specifically analyze and determine which dormancy genes play an important role in the development and prognosis of NB. Since no single marker can represent the effect of dormancy, and a single gene can be modulated by various factors that cause an inaccurate predictive effect, a gene signature comprising various genes is recommended. Based on dormancy-associated gene signatures, the combination of multiple genes could increase the safety of posttreatment active surveillance and more accurately reflect the prognosis of NB. Nomograms integrate multiple key factors to evaluate prognosis, including molecular biology and clinicopathological parameters. They have considerable clinical significance in research and are widely used in oncology.
In this study, we downloaded NB gene expression data and clinical information from the Gene Expression Omnibus and ArrayExpress databases. Based on consensus clustering of the expression data of dormancy-associated genes, the NB samples were divided into different groups, and DEGs were explored between groups. Functional analyses of DEGs were performed, followed by the assessment of tumor immunity and the establishment of a predictive dormancy signature for NB patients. Cox regression analysis was then performed to identify and optimize the combination of 6 dormancy-associated genes (CDKN2A, also known as p16; BHLHB3, known as DEC2; CDKN2B, known as p15; MAPK14, known as p38; CDKN1B, known as p27; and BMP7) with prognostic value for NB rather than a single gene. Finally, gender, age, INSS stage, and MYCN status were identified as independent overall survival-related variables and incorporated into the nomogram. According to our findings, the nomogram is a reliable tool for NB clinical diagnosis and therapy evaluation. These results indicated that the 6-gene signature had a powerful capacity to predict NB prognosis, which had certain guiding significance in decision-making for clinical treatment.
Among the 6 genes, BMP7 has been found to influence many human solid carcinomas, including colorectal, ovarian, breast, prostate, and cervical cancers, and can predict disease aggression and poor prognosis. Bone morphogenetic proteins (BMPs) are reported to be essential for development and differentiation of the nervous system and regulating cytoskeletal remodeling during neuronal morphogenesis (19). Despite the limited report, BMP7 may indirectly promote the cell proliferation and viability (20). Particularly for prostate cancer, higher expression level of BMP7 is significantly correlated with chemotherapy-resistance, recurrent metastatic disease, and stemness, which are hallmarks of dormant cancer cells (21). Kobayashi et al. systematically showed that BMP7 could induce dormancy in prostate cancer stem-like cells by activating MAPK14 and increasing expression levels of the cell cycle inhibitor p21 and the metastasis suppressor gene NDRG1 (N-myc downstream-regulated gene 1), indicating the influence of the BMP7-MAPK14-NDRG1 axis on dormancy and recurrence in prostate cancer (22).
BHLHB3, transcriptional repressor of E-box activity, is expressed in various embryonic and adult tissues and functions as a tumor suppressor, however, the role of BHLHB3 in NB progression remain unclear. Mechanistically, a potential role of BHLHB3 in regulating cell proliferation and apoptosis has been proposed. Recent evidence has shown that in breast cancer and salivary adenoid cystic carcinoma, BHLHB3 participated in limiting tumor growth, maintaining cell population arrested in G0/G1 phase, and induced tumor dormancy, which promotes tumor development and relapse (23,24).
As a potential regulatory target, activation of MAPK14 has many important physiological functions, for instance, it may provide a dual regulation of cell survival maintenance and apoptosis promotion in NB (25,26). Published evidence has shown that depending on the stimulating signal, increased MAPK14 activity could block cell proliferation by inhibiting activation of ERK in culture, which induces G0/G1 arrest or triggers senescence or apoptosis (27,28). Additional studies have demonstrated that a decreased ratio of ERK/p38 may induce disseminated tumor cells to enter a state of prolonged dormancy and that the ERK/p38 activity balance serves as a valid general predictor of dormancy in vivo (27,29). Simply put, ERK activity is negatively regulated by p38, and a high ERK/p38 ratio favors tumor cell proliferation, while a lower p38/ERK ratio prompts tumor dormancy. Moreover, the p38/ERK ratio can also be regulated by urokinase plasminogen activator receptor (uPAR) or fibronectin, which in turn influences tumor cell proliferation or dormancy in head and neck carcinoma (30).
Cell cycle regulators play critical roles in the equilibrium between cell dormancy and proliferation. The levels of cell cycle progression genes in quiescent cells are often discussed. Oncogene inactivation can prevent self-renewal of cancer cells and cause a conversion of tumor cell proliferation to reversible dormancy through induction of the expression of cell cycle arrest proteins (31). The INK4 family (e.g, p15, p16) and the Cip/Kip family (e.g, p27) of CDK inhibitors have been reported to be critical for cell proliferation and quiescence maintenance. The role of them in NB development functions differently. In some studies, upregulation of proliferative activity in NB cells was found to be accompanied by a decrease of cell cycle inhibitors (p15 and p27) while elevated levels of p27 may play a key role in RA induced growth arrest and neuronal differentiation of NB cells. Furthermore, abnormal expression of the p16 was associated with a poor prognosis in NB (32,33). As described previously, the CDK inhibitor CDKN1B is often highly expressed in quiescent cells, while the expression of proliferation marker MKI67 is low. Moreover, CDKN1B and Mki67 expression levels change inversely and progressively as cells exit and re-enter the cell cycle. Therefore, the combination of high CDKN1B and low MKI67 could serve as a marker of quiescent cancer cells (34). Zou et al. reported that CDKN1B had broad antiproliferative effects on a variety of tumor cell types, and that CDKN1B was a key intracellular mechanism in controlling hematopoietic stem cell dormancy (35). Van Delft found that in B-cell precursor childhood acute lymphoblastic leukemia, deletions of CDKN2A/B increased from 38% at presentation to 76% in relapse, further suggesting that cell-cycle deregulation contributed to extended periods of dormancy and emergence of relapse.
Due to tumor dormancy, most high-risk NBs present with widespread metastatic disease at diagnosis and either do not respond to conventional therapies initially or ultimately relapse after treatment. But each of these individual gene mentioned above can be modulated by various factors, a gene signature comprising various genes is recommended. In this study, a dormancy-associated gene signature, including CDKN2A, BHLHB3, CDKN2B, MAPK14, CDKN1B, and BMP7, was established. The gene signature showed a strong capacity to predict NB patient prognosis and had certain guiding significance in decision-making for clinical treatment. Through the nomogram, physicians might predict the overall survival more accurately and offer a reasonable personalized therapy for improving the survival of NB patients. Upon further clinical validation, patients classified as high-risk could receive more attention and intensive treatment, whereas excessive treatment should be avoided for those classified as low-risk.
Although the present study is the first to incorporate a dormancy-associated signature for predicting NB survival, it had some limitations. First, detailed clinicopathological information downloaded from the GSE database, such as operation extent and chemoradiation, were limited and incomplete and not included in the nomogram. Second, some of the molecules in the 6-gene signature and nomogram model have been confirmed. For example, higher expression of CDKN2A and BMP7 was found in tumor tissues of the late recurrence group in line with previous studies (22,32,33). However, more confirmatory and prospective studies are necessary for further validation of the prediction nomogram.
Conclusions
Nomogram prognostic models that predict NB prognosis in children may facilitate better treatment decisions and assessments. The results of our present study suggested a 6-gene genetic signature based on dormancy could predict the prognosis of neuroblastoma patients and their response to immunotherapy.
Acknowledgments
We would like to acknowledge the GEO and ArrayExpress networks for providing the gene expression data.
Funding: This work was supported by grants from the National Nature Science Foundation of China (Nos. 81903055, 81902401, and 82103672), Natural Science Foundation of Tianjin (No. 21JCQNJC01290), Tianjin Health Science and Technology Project (No. TJWJ2022MS008), National Key R&D Program of China (No. 2018YFC1313000), Tianjin Key Medical Discipline (Specialty) Construction Project (No. TJYXZDXK-009A).
Footnote
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-23-119/rc
Data Sharing Statement: Available at https://tp.amegroups.com/article/view/10.21037/tp-23-119/dss
Peer Review File: Available at https://tp.amegroups.com/article/view/10.21037/tp-23-119/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-23-119/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the by the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital (No. E20210027). Written informed consent was obtained from the parents or legal guardians of each patient.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Zafar A, Wang W, Liu G, et al. Molecular targeting therapies for neuroblastoma: Progress and challenges. Med Res Rev 2021;41:961-1021. [Crossref] [PubMed]
- Irwin MS, Naranjo A, Zhang FF, et al. Revised Neuroblastoma Risk Classification System: A Report From the Children’s Oncology Group. J Clin Oncol 2021;39:3229-41. [Crossref] [PubMed]
- Park JR, Kreissman SG, London WB, et al. Effect of Tandem Autologous Stem Cell Transplant vs Single Transplant on Event-Free Survival in Patients With High-Risk Neuroblastoma: A Randomized Clinical Trial. JAMA 2019;322:746-55. [Crossref] [PubMed]
- Twist CJ, Schmidt ML, Naranjo A, et al. Maintaining Outstanding Outcomes Using Response- and Biology-Based Therapy for Intermediate-Risk Neuroblastoma: A Report From the Children’s Oncology Group Study ANBL0531. J Clin Oncol 2019;37:3243-55. [Crossref] [PubMed]
- Tang J, Lu H, Yang Z, et al. Associations between WTAP gene polymorphisms and neuroblastoma susceptibility in Chinese children. Transl Pediatr 2021;10:146-52. [Crossref] [PubMed]
- Park JA, Cheung NV. Targets and Antibody Formats for Immunotherapy of Neuroblastoma. J Clin Oncol 2020;38:1836-48. [Crossref] [PubMed]
- Kushner BH, Helson L, Lane JM, et al. Metastatic neuroblastoma after 52 years of apparent dormancy. N Engl J Med 1986;315:196-7. [Crossref] [PubMed]
- Phan TG, Croucher PI. The dormant cancer cell life cycle. Nat Rev Cancer 2020;20:398-411. [Crossref] [PubMed]
- Recasens A, Munoz L. Targeting Cancer Cell Dormancy. Trends Pharmacol Sci 2019;40:128-41. [Crossref] [PubMed]
- Zhang W, Yu Y, Hertwig F, et al. Comparison of RNA-seq and microarray-based models for clinical endpoint prediction. Genome Biol 2015;16:133. [Crossref] [PubMed]
- Roderwieser A, Sand F, Walter E, et al. Telomerase Is a Prognostic Marker of Poor Outcome and a Therapeutic Target in Neuroblastoma. JCO Precis Oncol 2019;3:1-20. [Crossref] [PubMed]
- Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 2010;26:1572-3. [Crossref] [PubMed]
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun 2013;4:2612. [Crossref] [PubMed]
- Becht E, Giraldo NA, Lacroix L, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol 2016;17:218. [Crossref] [PubMed]
- Laut AK, Dorneburg C, Fürstberger A, et al. CHD5 inhibits metastasis of neuroblastoma. Oncogene 2022;41:622-33. [Crossref] [PubMed]
- Nakata R, Shimada H, Fernandez GE, et al. Contribution of neuroblastoma-derived exosomes to the production of pro-tumorigenic signals by bone marrow mesenchymal stromal cells. J Extracell Vesicles 2017;6:1332941. [Crossref] [PubMed]
- Eixarch H, Calvo-Barreiro L, Montalban X, et al. Bone morphogenetic proteins in multiple sclerosis: Role in neuroinflammation. Brain Behav Immun 2018;68:1-10. [Crossref] [PubMed]
- Wilkemeyer MF, Sebastian AB, Smith SA, et al. Antagonists of alcohol inhibition of cell adhesion. Proc Natl Acad Sci U S A 2000;97:3690-5. [Crossref] [PubMed]
- Cackowski FC, Heath EI. Prostate cancer dormancy and recurrence. Cancer Lett 2022;524:103-8. [Crossref] [PubMed]
- Kobayashi A, Okuda H, Xing F, et al. Bone morphogenetic protein 7 in dormancy and metastasis of prostate cancer stem-like cells in bone. J Exp Med 2011;208:2641-55. [Crossref] [PubMed]
- Adam AP, George A, Schewe D, et al. Computational identification of a p38SAPK-regulated transcription factor network required for tumor cell quiescence. Cancer Res 2009;69:5664-72. [Crossref] [PubMed]
- Yang X, Wu JS, Li M, et al. Inhibition of DEC2 is necessary for exiting cell dormancy in salivary adenoid cystic carcinoma. J Exp Clin Cancer Res 2021;40:169. [Crossref] [PubMed]
- Guo F, He XB, Li S, Le W. A Central Role for Phosphorylated p38α in Linking Proteasome Inhibition-Induced Apoptosis and Autophagy. Mol Neurobiol 2017;54:7597-609. [Crossref] [PubMed]
- Stefàno E, Muscella A, Benedetti M, et al. Antitumor and antimigration effects of a new Pt compound on neuroblastoma cells. Biochem Pharmacol 2022;202:115124. [Crossref] [PubMed]
- Farino Reyes CJ, Pradhan S, Slater JH. The Influence of Ligand Density and Degradability on Hydrogel Induced Breast Cancer Dormancy and Reactivation. Adv Healthc Mater 2021;10:e2002227. [Crossref] [PubMed]
- Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer 2007;7:834-46. [Crossref] [PubMed]
- Aguirre-Ghiso JA, Estrada Y, Liu D, et al. ERK(MAPK) activity as a determinant of tumor growth and dormancy; regulation by p38(SAPK). Cancer Res 2003;63:1684-95. [PubMed]
- Cabarcas SM, Mathews LA, Farrar WL. The cancer stem cell niche--there goes the neighborhood? Int J Cancer 2011;129:2315-27. [Crossref] [PubMed]
- Bellovin DI, Das B, Felsher DW. Tumor dormancy, oncogene addiction, cellular senescence, and self-renewal programs. Adv Exp Med Biol 2013;734:91-107. [Crossref] [PubMed]
- Shen X, Xu X, Xie C, et al. YAP promotes the proliferation of neuroblastoma cells through decreasing the nuclear location of p27(Kip1) mediated by Akt. Cell Prolif 2020;53:e12734. [Crossref] [PubMed]
- Obana K, Yang HW, Piao HY, et al. Aberrations of p16INK4A, p14ARF and p15INK4B genes in pediatric solid tumors. Int J Oncol 2003;23:1151-7. [Crossref] [PubMed]
- Miller I, Min M, Yang C, et al. Ki67 is a Graded Rather than a Binary Marker of Proliferation versus Quiescence. Cell Rep 2018;24:1105-1112.e5. [Crossref] [PubMed]
- Zou P, Yoshihara H, Hosokawa K, et al. p57(Kip2) and p27(Kip1) cooperate to maintain hematopoietic stem cell quiescence through interactions with Hsc70. Cell Stem Cell 2011;9:247-61. [Crossref] [PubMed]


