The third generation sequencing: the advanced approach to genetic diseases
Introduction
The definition of a rare disease is a disease affecting fewer than 2,000 people in Europe (1), while, a disease affecting less than 200,000 people is defined as a rare disease in the United States. Though the chance that individuals who are diagnosed with each rare disease is seemingly low, approximately 7,000–8,000 rare diseases are estimated to date. The Global Genes Project estimates that 300 million people worldwide are affected by a rare disease and eighty percent of rare diseases are gene of origin (2). Moreover, about fifty percent of those affected by rare diseases are in their childhood, thirty percent of who will not survive beyond their fifth year old.
The current genomic technological advancement has changed the research approaches and clinical strategies of rare diseases. In the past few years, the human genome project was firstly completed in mapping all the genes in human with a cost of almost 3 billion dollars. Then the sequencing price significantly dropped with the application of the next generation sequencing (NGS). High throughput and low cost of genomic sequencing make the further insight into genetic diseases in more patients possible. However, the percentage of all known rare diseases with the pathogenic gene is less than fifty percent (3). One reason is that the routine sequencing technology has missed some mutations. Hence, it has still been a challenge to develop diagnostics, managements and genetic advice for these patients in practice.
This paper aims to provide a review of the third generation sequencing (TGS) in genetic diseases. A brief review of revolution of the sequencing technology in genetic diseases is firstly presented. And then, an overview of the TGS approach to the genetic diseases and its clinical effect follows. The bioinformatic methods applied to the new technology and limitations of the TGS are also discussed.
Brief revolution of sequencing technology
Aside from the study of genetic diseases via karyotyping, DNA microarrays, FISH or multiples ligation-dependent probe amplification (MLPA), the first generation sequencing, including Maxam Gilbert methods and Sanger sequencing opened up the new door into the genetic diseases since 1977.
The next generation sequencing (NGS) emerged in 2004 helped researchers gain deeper understanding about the genetic diseases (4). More than 2,400 pathogenic genes have been identified (5) and over 150 genetic diseases have been identified via the whole exome sequencing (WES) (6). Three important centers for Mendelian Genomics (CMGs) funded by NIH, including University of Washington, Yale University and the Baylor College of Medicine utilized the NGS to elucidate many Mendelian disorders (7). In short, the NGS has a great impact on de novo mutations in rare diseases in recent years. However, many rare diseases are still not fully diagnosed by the NGS due to the short-read methods (~150–300 bp). Structural variants (SVs), repetitive elements, extreme guanine-cytosine (GC) content or sequences with multiple homologous elements in the genome are difficult to be characterized via the NGS, even with the use of state-of-the-art bioinformatic algorithms (8,9). These drawbacks of the NGS-based investigations of human diseases have strongly driven the search for other methods to improve the accuracy and reduce diagnosing time in genetic diseases.
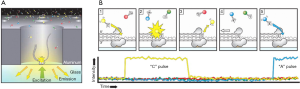
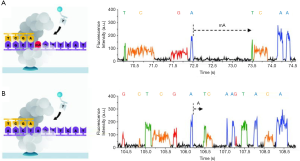
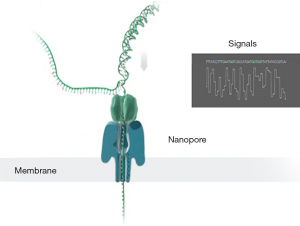
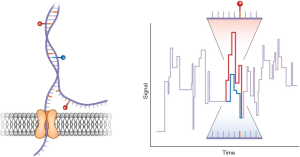
The TGS provided by Pacific Biosciences (PacBio) and Oxford Nanopore Technologies (ONT) in 2011, is a single molecular and real-time sequencing technology (10).The PacBio platform adopts single-molecule real-time (SMRT) technology (Figure 1). In the DNA library preparation, no PCR is required as a closed and circular ssDNA template can be replicated automatically. During the sequencing process, the fluorescence signals are activated by a laser as soon as a labeled dNTPs is incorporated into DNA. A camera system then records the color and duration of the emitted light in real time in the flow cell equipped with zero mode waveguides (ZMVs). The time of the base incorporation is longer as the base is modificated. Thus, the time called “interpulse duration” can indicate the DNA modification event (Figure 2) (11). The SMRT technology also allows the direct RNA-sequencing (12). The SMRT essentially is still based on sequencing by synthesis. Nanopore Sequencing Technology (ONT) utilizes nanopore inserted in an electrical resistant membrane. A potential is applied across the membrane, resulting in a current flowing only through the nanopore (Figure 3). The characteristic disruptions in the current can be measured, indicating a specific single molecular. In the DNA library preparation, a hairpin structure is designed to ligate the double DNA strands so that the system can read both DNA strands in one continuous read. As dsDNA moves through the nanopore, the bound polymerase or helicase enzyme can attach the DNA in the pore. During sequencing process, a characteristic disruption in the electrical current can be measured as the nucleotide passing through the nanopore (Figure 4) (13). Then the nucleotide can be identified. These features allow the detection of hundreds of kilobases in one continuous read. Ultra-long reads (ULRs) with above 300 kb reads and some close to 1 million bp reads can be sequenced in the ONT (14). Also, the many pocket-sized sequencers developed by the ONT are portable without sophisticated laboratory setup and can be transported out of the lab with low cost. For example, the MinION was transported to Africa for screening Ebola and Lassa virus outbreak (15,16). In short, the features of the TGS introduced by PacBio and ONT allow for the long-read sequencing in real-time with the low alignment and mapping errors during library construction.
Comprehensive genetic disease identification
The genetic disease can be understood in molecular level rather than that of chromosome owing to the development of sequencing technologies. The TGS has not only helped to discover more novel genetic diseases (17), but also revised the identification of genetic disease (17).
SV has an important role in genetic disorders (18). SVs are defined as mutations affecting more than 50 base pairs. The SVs include deletions, insertions, inversions, mobile element transpositions, translocations, tandem repeats and copy number variants (CNVs) (19). By using SMRT sequencing for two haploid human genomes, Huddleston’s group pointed out that estimated approximately 89% SVs have been missed in the 1,000 Genomes Project (20). Although the sophisticated SV genotyping software methods were available, the detection of SVs was low (30–70%) and the error rates were still high (85%) (21). The single molecular and real-time sequencing has shown a better capacity to discover the structural-variant events. A few SVs related genetic diseases detected through the TGS is reviewed in the following part. For example, Aneichyk and colleagues studied X-linked Dystonia-Parkinsonism (XDP) which is a Mendelian neurodegenerative disease and suggested that a SINE-WNTR-Alu (SVA) mediated aberrant transcriptional mechanism was associated with XDP (22). The precise breakpoints of the deletion in a homozygous 7p14.3 were deciphered in the proband with Barde-Biedl syndrome (BBS) and carrier parents by long-read SMRT sequencing (23). The WES yielded only one heterozygous causal variants in the patient with glycogen storage disease type Ia (GSD-Ia) which is an autosomal recessive disease (24), while, a 7.1 kb deletion covering two exons in G6PC on the other allele were detected through Nanopore long-read whole genome sequencing (WGS) (24). Multiple neoplasia and cardiac myxoma with the negative NGS results were found in connection with a heterozygous 2,184 bp deletion of the first coding exon of PRKAR1A (25). A complex novel translocation t(X;20)(q11.1;p13) was delineated via Nanopore long read sequencing (LRS) technology in a balanced reciprocal translocation (BRT) case (26). Other congenital diseases associated with complex chromothripsis were identified to link to the de novo complex SV breakpoints via ONT (27). In addition, fine-mapping of dipeptidyl-peptidase 6 gene (DPP6) in an autosomal dominant dementia family significantly linked to 7q36 was identified via the PromethION sequencing platform (Oxford Nanopore Technologies) (28).
Another advantage of the TGS is characterize the characterization of complete repeat expansion of genes and discriminate pseudogenes. As a typical example, the C9ofr72'GGGGCC' (G4C2) repeat expansion associated with amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) was validated through Pacific Biosciences and Oxford Nanopore Technologies (29). The CGG short tandem repeats in fragile X syndrome were detected by SMRT sequencing (30). Likely, the familial myoclonic epilepsy was connected with a 4.6 kb repeat expansion and 12.4 kb deletion in complex repeat regions via SMRT (31,32). Additionally, repeat expansions of complex genes, such as ATXN10, HTT, SMAD12, TNRC6A and RAPGEF2 were also validated by SMRT sequencing (33-36). CTG-repeat expansion was confirmed by SMRT in CRISPR/Cas9-mediated editing in myotonic dystrophy patient as well (37). Other complex and challenging regions of the human genome were characterized via the TGS, such as autosomal-dominant polycystic kidney disease (ADPKD). Duplicated and high GC content genomic regions as well as six pseudogenes of PKD1 gene can lead to ambiguous identification of variants via the NGS. However, 94.7% of the patients with PKD1 pathogenic variants were identified via SMAT by Borras and colleagues (38). A GC-rich 60 basepair variable number of tandem repeat (VNTR) and all variants position of the Mucin-1 gene in autosomal dominant tubulointerstitial kidney disease (ADTKD) were also determined by SMRT sequencing (39). Another example is the primary immunodeficiency-associated gene IKBKG. The pseudogene IKBKGP1 can be bypassed by long read, single-molecule sequencing which allows the rapid and efficient identification of the primary immunodeficiency diseases (40). Sanna Gudmundsson and colleagues clarified the mechanism of revertant mosaicism through SMRT (41). They demonstrated that the dominant negative effects of the p.Asp50Asn mutation was reverted by the second-site mutations of Cx25-Asp50Asn resulting in the development of healthy-looking skin in a patient with ichthyosis-deafness (KID) syndrome.
As stated above, the features of the TGS also allows the detection of the epigenetic modification in real time. DNA modifications have been found in a wide range of living organisms, from prokaryotes to eukaryotes. Many existing studies have shown that they play important roles in development diseases, such as lysosomal storage disorders, tumorigenesis, autoinflammatory diseases, imprinting and X chromosome inactivation (42-45). The bisulfite Sanger sequencing and other next generation sequencing have the restriction to read length of only 150–130 bp (46,47). Therefore, long-read single-molecule real-time bisulfite sequencing (SMRT-BS) developed by Yang and colleagues is a technique that combines bisulfite conversion with the TGS and allowed the detection of the targeted CpG methylation in real time (48).
Furthermore, LRS allows the detection of full-length mRNA transcript in one read. The short-read RNA sequencing always leads to inaccurate annotation due to computational transcript reconstruction (49). Aneichyk and colleagues utilized the long-read RNA-sequencing to decipher the TAF1 expression in the X-linked dystonia-parkinsonism (XDP) (22). Roeck and colleagues demonstrated that the Alzheimer’s disease severity was in relation to the varying degrees of nonsense-mediated mRNA decay (NMD) and transcript-modifying events (50). Twenty-seven genetically unsolved patients with an external collagen VI-like dystrophy were found in connection with highly recurrent de novo intronic mutation in COL6A1 via RNA-sequencing (51).
The combination of the TGS with other technologies, such as the NGS or single cell sequencing or target genome editing, will also give an insight into the genome and bring the new therapies (52). Mimori and colleagues utilized the SMRT sequencing and additional short-read data to obtain the high-quality and full-length human leukocyte antigen alleles reconstruction successfully (53). A study of Hendel A and colleagues showed that SMRT sequencing was facilitated to quantify the genome editing outcomes after the large genes were inserted at the endogenous IL2RG, HBB, and CCR5 loci by transcription activator-like effector nucleases (TALENs), zinc finger nucleases (ZFNs) or clustered regularly interspaced short palindromic repeats (CRISPR/Cas9 or RNA-guided endonucleases (RGENs) (54).
Clinical effect of the TGS
Importantly, clinical decisions and outcomes can be benefited from the TGS applications with more complete detection of mutations. For example, de novo mutations can occur in the different stage of embryonic development. Depending on the different stage of postzygotic mutation during development, such mutations may lead to somatic or germline mosaicism or both (55). Understanding of the complex SVs guided the genetic counseling and enable a successful preimplantation genetic diagnosis in the family (56). Maria and colleagues demonstrated that less than 1% of the TCOF1 variant c.3156C>T cells were in the paternal germ cells in a family with a child suffering Treacher Collins syndrome, suggesting the low recurrence risk in offspring (55). Similarly, there were 40% of PTPN11 variant c.923A>C cells in the paternal germ cells in a family with unsuccessful pregnancies, indicating a high recurrence risk of Noonan syndrome in offspring (55). Moreover, AGG interruptions in females with a FMR1 premutation were detected by long-read single-molecule sequencing, which was previously undetected due to the technical difficulties (57). In short, apart from the increasing discovery of novel disease genes, the TGS aids the preimplantation genetic counseling.
Ethics is also important in gene sequencing technology. The informed consent, data privacy and return of results are three issues demanding attention (58). To date, the recommendations of ethical considerations have been addressed by the American College of Medical Genetics and Genomics (ACMG) (59). Obviously, more ethical issues await the TGS as more discoveries of the novel disease genes in clinical practice come up.
Bioinformatic methods in the TGS
With more discoveries of novel SVs, repeat expansions and long noncoding RNAs (IncRNAs) via the TGS, the bioinformatic algorithms have to be TGS-specific and more user-friendly. The major bioinformatic challenges of the TGS is the high sequencing error rate which is 10–15% in the PacBio and 5–20% in the ONT. Therefore, the new alignment and error correction algorithms are required (Table 1). Several studies have offered the relatively new methods to correct the sequencing errors in the TGS. The methods for alignment and phasing are LAST (77), BLASR (73), BWA-MEM (74), GraphMap (75), MECAT (64) and minimap2 (78), PBHoney (79), NGMLR (74), Sniffles (74), CORGi (83), SIVM (84), SMRT-SV (95), NextSV (96), NanoSV (97) and Picky (98) in de novo mutations and SVs detection. Regarding the RNA sequencing analysis, the available bioinformatic tools include SQANTI (85), TAPIS (86), ToFU (87), BLAT (88), Gmap (89). In terms of errors correction in sequencing analysis, there are a few available methods as well. Hybrid error correction methods including Nanocorr, MaSCA, PBcR or Spades utilize short-read data to correct the error. However, because of biases in short-read coverage and repetitive sequence, FALCON-sense, HGAP, pbCR, Canu or MARVEL is more accurate as they are self-error correction methods (99). The other technique developed by Jana Ebler and colleagues combined the inference of haplotype and genotypes from noisy long reads (100). A similar software named as NanoSim is a fast and large-scale read simulator to call reads errors in MinION platforms (101). A TGS tool developed by Danze Chen’s group is a bioinformatic suit to compare isoforms, identify alternative splicing pattern and IncRNA (102). Moreover, a time and resource effective strategy for completing short read assembles has been applied, which enable sufficient analysis date to be assembled with the shortest sequencing time (103).

Full table
The TGS comes with several limitations (14,43,44). First, the DNA library required fresh material or intact cells and the protocols for the handing of ultra-long high molecular weight DNA require improvements. Second, the TGS has the challenges with the higher sequencing error rate and systematic error. Third, the cost of the TGS still has been higher than that of the NGS ($65–$200 per Gb in the PacBio and $22–$90 per Gb in the ONT). Additionally, because the database systems for interpreting complicated SVs are rare, thus the bioinformatic analysis are challenging.
Currently, the NGS is still our first choice of diagnosing the genetic diseases in clinical settings and the TGS can play a complementary role as a result of its limitations. However, with the maturation of the TGS approach, it will be widely used in researches and clinical practice. In the future, the picture of human genome will be more comprehensive as the more genomic data generated.
Acknowledgments
Thanks for Xinran Dong for the critical reading of the “Bioinformatic methods in the TGS” part of the manuscript.
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tp.2020.03.06). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Thorat C, Xu K, Freeman SN, et al. What the Orphan Drug Act Has Done Lately for Children With Rare Diseases: A 10-Year Analysis. Pediatrics 2012;129:516-21. [Crossref] [PubMed]
- Danielsson K, Mun LJ, Lordemann A, et al. Next-generation sequencing applied to rare diseases genomics. Expert Rev Mol Diagn 2014;14:469-87. [Crossref] [PubMed]
- Bacchelli C, Williams HJ. Opportunities and technical challenges in next-generation sequencing for diagnosis of rare pediatric diseases. Expert Rev Mol Diagn 2016;16:1073-82. [Crossref] [PubMed]
- Need AC, Shashi V, Hitomi Y, et al. Clinical application of exome sequencing in undiagnosed genetic conditions. J Med Genet 2012;49:353-61. [Crossref] [PubMed]
- Sobreira NL, Cirulli ET, Avramopoulos D, et al. Whole-genome sequencing of a single proband together with linkage analysis identifies a Mendelian disease gene. PLoS Genet 2010;6:e1000991. [Crossref] [PubMed]
- Rabbani B, Tekin M, Mahdieh N. The promise of whole-exome sequencing in medical genetics. J Hum Genet 2014;59:5-15. [Crossref] [PubMed]
- Ng SB, Bigham AW, Buckingham KJ, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet 2010;42:790-3. [Crossref] [PubMed]
- Salzberg SL, Yorke JA. Beware of mis-assembled genomes. Bioinformatics 2005;21:4320-1. [Crossref] [PubMed]
- Treangen TJ, Salzberg SL. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat Rev Genet 2011;13:36-46. [Crossref] [PubMed]
- van Dijk EL, Jaszczyszyn Y, Naquin D, et al. The Third Revolution in Sequencing Technology. Trends Genet 2018;34:666-81. [Crossref] [PubMed]
- Flusberg BA, Webster DR, Lee JH, et al. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods 2010;7:461-5. [Crossref] [PubMed]
- Eid J, Fehr A, Gray J, et al. Real-Time DNA Sequencing from Single Polymerase Molecules. Science 2009;323:133-8. [Crossref] [PubMed]
- Schatz MC. Nanopore sequencing meets epigenetics. Nat Methods 2017;14:347-8. [Crossref] [PubMed]
- Midha MK, Wu M, Chiu KP. Long-read sequencing in deciphering human genetics to a greater depth. Hum Genet 2019;138:1201-15. [Crossref] [PubMed]
- Kafetzopoulou LE, Pullan ST, Lemey P, et al. Metagenomic sequencing at the epicenter of the Nigeria 2018 Lassa fever outbreak. Science 2019;363:74. [Crossref] [PubMed]
- Hoenen T, Groseth A, Rosenke K, et al. Nanopore Sequencing as a Rapidly Deployable Ebola Outbreak Tool. Emerging Infectious Diseases 2016;22:331-4. [Crossref] [PubMed]
- Mantere T, Kersten S, Hoischen A. Long-Read Sequencing Emerging in Medical Genetics. Front Genet 2019;10:426. [Crossref] [PubMed]
- Sebat J, Lakshmi B, Troge J, et al. Large-scale copy number polymorphism in the human genome. Science 2004;305:525-8. [Crossref] [PubMed]
- Sudmant PH, Rausch T, Gardner EJ, et al. An integrated map of structural variation in 2,504 human genomes. Nature 2015;526:75-81. [Crossref] [PubMed]
- Huddleston J, Chaisson MJP, Steinberg KM, et al. Discovery and genotyping of structural variation from long-read haploid genome sequence data. Genome Res 2017;27:677-85. [Crossref] [PubMed]
- Chander V, Gibbs RA, Sedlazeck FJ. Evaluation of computational genotyping of structural variation for clinical diagnoses. Gigascience 2019;8:giz110. [Crossref] [PubMed]
- Aneichyk T, Hendricks WT, Yadav R, et al. Dissecting the causal mechanism of X-linked dystonia-parkinsonism by integrating genome and transcriptome assembly. Cell 2018;172:897 -909.e21. [PubMed]
- Reiner J, Pisani L, Qiao WQ, et al. Cytogenomic identification and long-read single molecule real-time (SMRT) sequencing of a Bardet-Biedl Syndrome 9 (BBS9) deletion. NPJ Genom Med 2018;3:3. [Crossref] [PubMed]
- Miao H, Zhou JP, Yang Q, et al. Long-read sequencing identified a causal structural variant in an exome-negative case and enabled preimplantation genetic diagnosis. Hereditas 2018;155:32. [Crossref] [PubMed]
- Merker JD, Wenger AM, Sneddon T, et al. Long-read genome sequencing identifies causal structural variation in a Mendelian disease. Genet Med 2018;20:159-63. [Crossref] [PubMed]
- Dutta UR, Rao SN, Pidugu VK, et al. Breakpoint mapping of a novel de novo translocation t(X;20)(q11.1;p13) by positional cloning and long read sequencing. Genomics 2019;111:1108-14. [Crossref] [PubMed]
- Cretu Stancu M, van Roosmalen MJ, Renkens I, et al. Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat Commun 2017;8:1326. [Crossref] [PubMed]
- Cacace R, Heeman B, Van Mossevelde S, et al. Loss of DPP6 in neurodegenerative dementia: a genetic player in the dysfunction of neuronal excitability. Acta Neuropathol 2019;137:901-18. [Crossref] [PubMed]
- Ebbert MTW, Farrugia SL, Sens JP, et al. Long-read sequencing across the C9orf72 'GGGGCC' repeat expansion: implications for clinical use and genetic discovery efforts in human disease. Mol Neurodegener 2018;13:46. [Crossref] [PubMed]
- Schmidt MHM, Pearson CE. Disease-associated repeat instability and mismatch repair. DNA Repair 2016;38:117-26. [Crossref] [PubMed]
- Mizuguchi T, Toyota T, Adachi H, et al. Detecting a long insertion variant in SAMD12 by SMRT sequencing: implications of long-read whole-genome sequencing for repeat expansion diseases. J Hum Genet 2019;64:191-7. [Crossref] [PubMed]
- Mizuguchi T, Suzuki T, Abe C, et al. A 12-kb structural variation in progressive myoclonic epilepsy was newly identified by long-read whole-genome sequencing. J Hum Genet 2019;64:359-68. [Crossref] [PubMed]
- Schüle B, McFarland KN, Lee K, et al. Parkinson's disease associated with pure ATXN10 repeat expansion. NPJ Parkinsons Dis 2017;3:27. [Crossref] [PubMed]
- Ishiura H, Doi K, Mitsui J, et al. Expansions of intronic TTTCA and TTTTA repeats in benign adult familial myoclonic epilepsy. Nat Genet 2018;50:581-90. [Crossref] [PubMed]
- Zeng S, Zhang MY, Wang XJ, et al. Long-read sequencing identified intronic repeat expansions in SAMD12 from Chinese pedigrees affected with familial cortical myoclonic tremor with epilepsy. J Med Genet 2019;56:265-70. [Crossref] [PubMed]
- Höijer I, Tsai YC, Clark TA, et al. Detailed analysis of HTT repeat elements in human blood using targeted amplification-free long-read sequencing. Hum Mutat 2018;39:1262-72. [Crossref] [PubMed]
- Dastidar S, Ardui S, Singh K, et al. Efficient CRISPR/Cas9-mediated editing of trinucleotide repeat expansion in myotonic dystrophy patient-derived iPS and myogenic cells. Nucleic Acids Res 2018;46:8275-98. [Crossref] [PubMed]
- Borràs DM, Vossen RHAM, Liem M, et al. Detecting PKD1 variants in polycystic kidney disease patients by single-molecule long-read sequencing. Hum Mutat 2017;38:870-9. [Crossref] [PubMed]
- Wenzel A, Altmueller J, Ekici AB, et al. Single molecule real time sequencing in ADTKD-MUC1 allows complete assembly of the VNTR and exact positioning of causative mutations. Sci Rep 2018;8:4170. [Crossref] [PubMed]
- Frans G, Meert W, Van der Werff Ten Bosch J, et al. Conventional and Single-Molecule Targeted Sequencing Method for Specific Variant Detection in IKBKG while Bypassing the IKBKGP1 Pseudogene. J Mol Diagn 2018;20:195-202. [Crossref] [PubMed]
- Gudmundsson S, Wilbe M, Ekvall S, et al. Revertant mosaicism repairs skin lesions in a patient with keratitis-ichthyosis-deafness syndrome by second-site mutations in connexin 26. Hum Mol Genet 2017;26:1070-7. [Crossref] [PubMed]
- Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet 2000;16:168-74. [Crossref] [PubMed]
- Mohandas T, Sparkes RS, Shapiro LJ. Reactivation of an inactive human X chromosome: evidence for X inactivation by DNA methylation. Science 1981;211:393-6. [Crossref] [PubMed]
- Álvarez-Errico D, Vento-Tormo R, Ballestar E. Genetic and Epigenetic Determinants in Autoinflammatory Diseases. Front Immunol 2017;8:318. [Crossref] [PubMed]
- Hassan S, Sidransky E, Tayebi N. The role of epigenetics in lysosomal storage disorders: Uncharted territory. Mol Genet Metab 2017;122:10-8. [Crossref] [PubMed]
- Taylor KH, Kramer RS, Davis JW, et al. Ultradeep bisulfite sequencing analysis of DNA methylation patterns in multiple gene promoters by 454 sequencing. Cancer Res 2007;67:8511-8. [Crossref] [PubMed]
- Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc 2007;2:2265-75. [Crossref] [PubMed]
- Yang Y, Sebra R, Pullman BS, et al. Quantitative and multiplexed DNA methylation analysis using long-read single-molecule real-time bisulfite sequencing (SMRT-BS). BMC Genomics 2015;16:350. [Crossref] [PubMed]
- Steijger T, Abril JF, Engstrom PG, et al. Assessment of transcript reconstruction methods for RNA-seq. Nat Methods 2013;10:1177-84. [Crossref] [PubMed]
- De Roeck A, Van den Bossche T, van der Zee J, et al. Deleterious ABCA7 mutations and transcript rescue mechanisms in early onset Alzheimer's disease. Acta Neuropathologica 2017;134:475-87. [Crossref] [PubMed]
- Cummings BB, Marshall JL, Tukiainen T, et al. Cummings BB, Marshall JL, Tukiainen T, et al. Improving genetic diagnosis in Mendelian disease with transcriptome sequencing. Sci Transl Med 2017;9:eaal5209.
- Hestand MS, Ameur A. The Versatility of SMRT Sequencing. Genes (Basel) 2019;10:24. [Crossref] [PubMed]
- Mimori T, Yasuda J, Kuroki Y, et al. Construction of full-length Japanese reference panel of class I HLA genes with single-molecule, real-time sequencing. Pharmacogenomics J 2019;19:136-46. [Crossref] [PubMed]
- Hendel A, Kildebeck EJ, Fine EJ, et al. Quantifying Genome-Editing Outcomes at Endogenous Loci with SMRT Sequencing. Cell Rep 2014;7:293-305. [Crossref] [PubMed]
- Wilbe M, Gudmundsson S, Johansson J, et al. A novel approach using long-read sequencing and ddPCR to investigate gonadal mosaicism and estimate recurrence risk in two families with developmental disorders. European Journal of Human Genetics 2019;27:849-50.
- Miao H, Zhou J, Yang Q, et al. Long-read sequencing identified a causal structural variant in an exome-negative case and enabled preimplantation genetic diagnosis. Hereditas 2018;155:32. [Crossref] [PubMed]
- Ardui S, Race V, de Ravel T, et al. Detecting AGG Interruptions in Females With a FMR1 Premutation by Long-Read Single-Molecule Sequencing: A 1 Year Clinical Experience. Front Genet 2018;9:150. [Crossref] [PubMed]
- Danielsson K, Mun LJ, Lordemann A, et al. Next-generation sequencing applied to rare diseases genomics. Expert Rev Mol Diagn 2014;14:469-87. [Crossref] [PubMed]
- Green RC, Berg JS, Grody WW, et al. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 2013;15:565-74. [Crossref] [PubMed]
- Berlin K, Koren S, Chin CS, et al. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat Biotechnol 2015;33:623-30. [Crossref] [PubMed]
- Li H. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 2016;32:2103-10. [Crossref] [PubMed]
- Koren S, Walenz BP, Berlin K, et al. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res 2017;27:722-36. [Crossref] [PubMed]
- Kamath GM, Shomorony I, Xia F, et al. HINGE: long-read assembly achieves optimal repeat resolution. Genome Res 2017;27:747-56. [Crossref] [PubMed]
- Xiao CL, Chen Y, Xie SQ, et al. MECAT: fast mapping, error correction, and de novo assembly for single-molecule sequencing reads. Nat Methods 2017;14:1072-4. [Crossref] [PubMed]
- Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012;19:455-77. [Crossref] [PubMed]
- Chin CS, Alexander DH, Marks P, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 2013;10:563-9. [Crossref] [PubMed]
- Lin Y, Yuan J, Kolmogorov M, et al. Assembly of long error-prone reads using de Bruijn graphs. Proc Natl Acad Sci U S A 2016;113:E8396-405. [Crossref] [PubMed]
- Nowoshilow S, Schloissnig S, Fei JF, et al. The axolotl genome and the evolution of key tissue formation regulators. Nature 2018;554:50-5. [Crossref] [PubMed]
- Warren RL, Yang C, Vandervalk BP, et al. LINKS: Scalable, alignment-free scaffolding of draft genomes with long reads. Gigascience 2015;4:35. [Crossref] [PubMed]
- Cao MD, Nguyen SH, Ganesamoorthy D, et al. Scaffolding and completing genome assemblies in real-time with nanopore sequencing. Nat Commun 2017;8:14515. [Crossref] [PubMed]
- English AC, Richards S, Han Y, et al. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS One 2012;7:e47768. [Crossref] [PubMed]
- Vaser R, Sovic I, Nagarajan N, et al. Fast and accurate de novo genome assembly from long uncorrected reads. Genome Res 2017;27:737-46. [Crossref] [PubMed]
- Chaisson MJ, Tesler G. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics 2012;13:238. [Crossref] [PubMed]
- Sedlazeck FJ, Rescheneder P, Smolka M, et al. Accurate detection of complex structural variations using single-molecule sequencing. Nat Methods 2018;15:461-8. [Crossref] [PubMed]
- Sović I, Sikic M, Wilm A, et al. Fast and sensitive mapping of nanopore sequencing reads with GraphMap. Nat Commun 2016;7:11307. [Crossref] [PubMed]
- Liu B, Gao Y, Wang Y. LAMSA: fast split read alignment with long approximate matches. Bioinformatics 2017;33:192-201. [Crossref] [PubMed]
- Kiełbasa SM, Wan R, Sato K, et al. Adaptive seeds tame genomic sequence comparison. Genome Res 2011;21:487-93. [Crossref] [PubMed]
- Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 2018;34:3094-100. [Crossref] [PubMed]
- English AC, Salerno WJ, Reid JG. PBHoney: identifying genomic variants via long-read discordance and interrupted mapping. BMC Bioinformatics 2014;15:180. [Crossref] [PubMed]
- Chin CS, Peluso P, Sedlazeck FJ, et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods 2016;13:1050-4. [Crossref] [PubMed]
- Edge P, Bafna V, Bansal V. HapCUT2: robust and accurate haplotype assembly for diverse sequencing technologies. Genome Res 2017;27:801-12. [Crossref] [PubMed]
- Patterson M, Marschall T, Pisanti N, et al. WhatsHap: Weighted Haplotype Assembly for Future-Generation Sequencing Reads. J Comput Biol 2015;22:498-509. [Crossref] [PubMed]
- Stephens Z, Wang C, Iyer RK, et al. Detection and visualization of complex structural variants from long reads. BMC Bioinformatics 2018;19:508. [Crossref] [PubMed]
- Heller D, Vingron M. SVIM: structural variant identification using mapped long reads. Bioinformatics 2019;35:2907-15. [Crossref] [PubMed]
- Tardaguila M, de la Fuente L, Marti C, et al. SQANTI: extensive characterization of long-read transcript sequences for quality control in full-length transcriptome identification and quantification. Genome Res 2018;28:396-411. Erratum in: Genome Res 2018;28:1096. [Crossref] [PubMed]
- Abdel-Ghany SE, Hamilton M, Jacobi JL, et al. A survey of the sorghum transcriptome using single-molecule long reads. Nature Communications 2016;7:11706. [Crossref] [PubMed]
- Gordon SP, Tseng E, Salamov A, et al. Widespread Polycistronic Transcripts in Fungi Revealed by Single-Molecule mRNA Sequencing. PLoS One 2015;10:e0132628. [Crossref] [PubMed]
- Kent WJ. BLAT - The BLAST-like alignment tool. Genome Res 2002;12:656-64. [Crossref] [PubMed]
- Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 2005;21:1859-75. [Crossref] [PubMed]
- Simpson JT, Workman RE, Zuzarte PC, et al. Detecting DNA cytosine methylation using nanopore sequencing. Nat Methods 2017;14:407-10. [Crossref] [PubMed]
- Rand AC, Jain M, Eizenga JM, et al. Mapping DNA methylation with high-throughput nanopore sequencing. Nat Methods 2017;14:411-3. [Crossref] [PubMed]
- Goodwin S, Gurtowski J, Ethe-Sayers S, et al. Oxford Nanopore sequencing, hybrid error correction, and de novo assembly of a eukaryotic genome. Genome Res 2015;25:1750-6. [Crossref] [PubMed]
- Zimin AV, Marcais G, Puiu D, et al. The MaSuRCA genome assembler. Bioinformatics 2013;29:2669-77. [Crossref] [PubMed]
- Koren S, Schatz MC, Walenz BP, et al. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol 2012;30:693-700. [Crossref] [PubMed]
- Huddleston J, Chaisson MJP, Steinberg KM, et al. Discovery and genotyping of structural variation from long-read haploid genome sequence data. Genome Res. 2017;27:677-85. Erratum in: Genome Res 2018;28:144. [Crossref] [PubMed]
- Fang L, Hu J, Wang D, et al. NextSV: a meta-caller for structural variants from low-coverage long-read sequencing data. BMC Bioinformatics 2018;19:180. [Crossref] [PubMed]
- Cretu Stancu M, van Roosmalen MJ, Renkens I, et al. Mapping and phasing of structural variation in patient genomes using nanopore sequencing. Nat Commun 2017;8:1326. [Crossref] [PubMed]
- Gong L, Wong CH, Cheng WC, et al. Picky comprehensively detects high-resolution structural variants in nanopore long reads. Nat Methods 2018;15:455-60. [Crossref] [PubMed]
- Sedlazeck FJ, Lee H, Darby CA, et al. Piercing the dark matter: bioinformatics of long-range sequencing and mapping. Nat Rev Genet 2018;19:329-46. [Crossref] [PubMed]
- Ebler J, Haukness M, Pesout T, et al. Haplotype-aware diplotyping from noisy long reads. Genome Biology 2019;20:116. [Crossref] [PubMed]
- Yang C, Chu J, Warren RL, et al. NanoSim: nanopore sequence read simulator based on statistical characterization. Gigascience 2017;6:1-6. [Crossref] [PubMed]
- Chen D, Zhao Q, Jiang L, et al. TGStools: A Bioinformatics Suit to Facilitate Transcriptome Analysis of Long Reads from Third Generation Sequencing Platform. Genes (Basel) 2019. [Crossref] [PubMed]
- Cao MD, Nguyen SH, Ganesamoorthy D, et al. Scaffolding and completing genome assemblies in real-time with nanopore sequencing. Nat Commun 2017;8:14515. [Crossref] [PubMed]