
An intelligent prediagnosis system for disease prediction and examination recommendation based on electronic medical record and a medical-semantic-aware convolution neural network (MSCNN) for pediatric chronic cough
Introduction
Cough is the most common symptom leading to pediatric medical treatment (1). In nonspecialized pediatric medical institutions, the proportion of children with cough as the first chief complaint is more than 35% (2). Cough is considered chronic in children when it is present for longer than 4 weeks (3). Etiologies of pediatric chronic cough include asthma, bacterial pharyngitis, influenza (FLU), acute upper respiratory tract infection, suppurative tonsillitis, and bacterial bronchitis. The high phenotypic similarities between pediatric respiratory diseases make it challenging to accurately diagnose chronic cough.
Recent research on this subject has focused on the use of artificial intelligence (AI) to aid the diagnosis of respiratory diseases (4-8), especially the diagnosis of coronavirus disease 2019 (COVID-19). However, most studies investigated intelligent diagnosis assistance during the formal diagnostic phase when medical examinations have been performed. In the early stage of medical treatment (referred to as the “prediagnosis stage” in this paper), many challenges exist that have not yet been addressed by researchers.
In the prediagnosis stage, the doctor must conduct a detailed interview with the patient, make a preliminary judgment of the disease based on the chief complaints described by the patient, and determine a succeeding examination plan for accurate diagnosis and treatment. Imaging examinations like chest X-ray (CXR) or chest computed tomography (CT) and physical examinations like lung auscultation are common ways for diagnosis of respiratory disease. Nevertheless, the misuse of imaging examination is very common in the pediatric respiratory clinic, especially in primary-level hospitals (9). According to the guidelines of pediatric chronic cough (2), mild cases of pneumonia do not require imaging examination, and strict criteria are used to determine which patients require a CT scan. However, in practice, many doctors do not strictly follow the guidelines for examination, especially in countries with insufficient medical human resources. Some health workers who have not been rigorously trained in clinical diagnosis lack knowledge about the guidelines. The excessive use of imaging examinations may cause negative effects to children’s health (10). The incidence of cancer caused by ionizing radiation in people aged 0–19 years is 3 times higher than that in adults over 20 years of age (11). On the other side, inaccurate prediagnosis of disease and inadequate examinations are also harmful as they may prolong the disease, leading to disruptive symptoms and a greater economic burden to patients and their families. Hence, there is an urgent need for methods that can assist doctors in the prediagnosis stage to accurately predict diseases and recommend appropriate examinations to patients in pediatric respiratory clinics.
In this paper, we designed an intelligent method to assist doctors in disease prediction and examination recommendation based on the clinical information in electronic medical record (EMR) text during the prediagnosis stage. EMR clinical notes contain rich information gained in doctor-patient interview process, such as the patient’s age, chief complaint, present disease history, past history, family history, history of allergy, and medication history, etc. These information are essential and useful for disease identification and examination determination in prediagnosis stage. Usually, an experienced and senior physician can make an accurate preliminary disease prediction by looking at the details in the chief complaint description, such as sputum and nasal mucus, cough sound, fever condition, panting, lung auscultation, etc., which are lacked in low-level hospitals. So in this paper, we chose to leverage the diagnostic experience of advanced children's hospitals. We constructed a pre-diagnostic framework based on the retrospective EMR data to help physicians judge diseases and make examination plans.
Recently, as the progress of natural language processing (NLP) techniques and deep learning algorithm, more and more researchers started to use EMR data for disease diagnose. For example, Kam et al. (12) extracted EMR data of multiple biological signal variables from the MIMIC II database and built a prediction network using a DNN model to facilitate the early detection of sepsis. Chao et al. (13) also used EMR data combined with time-series data and a recurrent neural network (RNN) to predict Parkinson’s disease. Wang et al. (14) constructed a prediction model using EHRs and a knowledge-based CNN to estimate the distant recurrence probability of patients with breast cancer. However, there is still no research leveraging EMR data in prediagnosis stage. Accurately predicting disease in the prediagnosis stage is not easy, as there are limited modes of evidence. Besides, to accurately understand the medical semantics from unstructured chief complaint text is also a challenge problem. To the best of our knowledge, this is the first study to use AI methods to assist doctors in the prediagnosis stage.
To achieve our goal, we developed a MSCNN framework for disease prediction and examination recommendation. This framework consisted of a pretrained medical language model and 2 TextCNN-based models. We trained the medical language model with a medical literature corpus to achieve better semantic representation of medical textual contents. For disease prediction and examination recommendation, we used NLP technologies to process patient information in EMR text. Our system learned to predict disease and recommend examinations through 2 respective TextCNN networks derived from transfer learning from the pretrained language models. To evaluate our method, we compared its performance with 4 other methods and designed an AC indication for the examination recommendation task. Experimental results showed that our method achieved a better performance in disease prediction than the 4 other models and obtained a good performance in examination recommendation.
The main contribution of this work lies in the following three aspects. First, we addressed diagnostic challenges in the prediagnosis stage of medical treatment, which has been ignored in previous studies of AI diagnosis assistance. Second, we developed an intelligent system to help doctors predict diagnoses and make appropriate examination plans for respiratory diseases with chronic cough as the chief complaint. Third, we designed a MSCNN framework that generates a more accurate semantic representation for medical text and can be rapidly transferred to downstream AI tasks.
This paper is organized as follows. Section 2 reviews relevant literature. Section 3 presents our method in detail, including the problem definition, system framework, and models. Section 4 reports on the experiments for the 2 AI tasks (disease prediction and examination recommendation) and their results, including the experimental set, evaluation metrics, experimental results, and discussion. Finally, Section 5 discusses this research further and concludes the paper. We present the following article in accordance with the STARD reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-22-275/rc).
Methods
In this section, we first describe the overall framework of our method. Then, we introduce the formal definitions of prediagnostic disease prediction and examination recommendation. Last, we present the two models by describing their dataset, algorithm, and the statistical methods for their performance assessment.
Overall framework
In this paper, we developed a medical-semantic-aware framework (MSCNN) to predict diseases and recommend examinations in the prediagnosis stage. As shown in Figure 1, the overall framework consisted of 3 main parts: a pre-trained language model based on word2vec (15) algorithms that improves awareness of medical semantics in downstream tasks, a TextCNN-based disease prediction model, and a TextCNN-based examination recommendation model whose parameters were obtained from transfer learning from the pretrained language model. The pretrained language model generated vector representations with a deeper awareness of medical content to improve the semantic foundation of the downstream tasks. The word embeddings output of the pretrained language model was taken as the input for the succeeding tasks. For disease prediction and examination recommendation, we built 2 TextCNN models with EMR data. The configuration of the 2 models was acquired from the pretrained language model through fine-tuning.
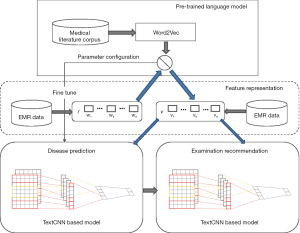
Problem definition
We formally define an EMR dataset with R medical records as . Each record r in set R can be seen as a sequence containing n words, denoted as , among which each word w is associated with a feature vector v according to the vocabulary. This vector corresponds to the word-embedding in the matrix , in which L denotes the scale of the embedding.
Definition for disease prediction
Assuming a collection of m different types of diseases that has the phenotype of chronic cough, given an EMR dataset X, the goal of the disease prediction task was as follows: for a chronic cough case, our prediagnostic prediction model will learn through the feature vectors extracted from EMR data and output the probability distribution of diseases for the case. Thus, the model will output a disease probability distribution vector , in which and . The higher the value of , the greater the incidence probability of the disease.
Definition for examination recommendation
Assuming a set composed of k types of examinations, denoted as , given an EMR dataset X, the goal of the examination recommendation task was as follows: for a chronic cough case and its disease prediction result d, the model will output an examination plan formatted as an examination distribution vector , in which . When equals 1, the model recommends taking a certain examination; when equals 0, the model does not recommend taking a certain examination.
Pretrained medical language model
Word embedding models based on a general corpus are insensitive in medical semantic contexts (16). In this work, we trained a medical language model using the word2vec (15) algorithm to obtain better semantic representations for medical-related content. Word2vec is a widely adopted model in NLP that maps high-dimensional one-hot word vectors to a low-dimensional dense word vector space for feature representation. Word2vec can be specifically divided into Skip-gram and continuous bag of words (CBOW) models (15). We applied the Skip-gram model to train the medical literature corpus, since the frequency of medical words is less than that of common words. By using central words to predict context words and making adjustments (17), Skip-gram is able to improve the accuracy of a semantic vector.
Given the text sequence [W1, W2, …, WT] and the word Wt as the center word, the probability of the context word will be maximized in the fixed window. With c denoting the size of the context window, the objective function of the algorithm is:
We calculated the probability in the objective function through the softmax function as follows:
Here WI represents the central word, W0 represents the context word, and VW and V’W represent the vectors of the input and output words, respectively. Each node of the softmax function outputs a value between 0 and 1, and the probability of all neuron nodes in the output layer sums to 1. To simplify the calculation of the softmax function, we leveraged the Negative Sampling (18) algorithm to selectively update part of the weights of the training sample to accelerate the gradient descent.
Disease prediction model
Dataset
For disease prediction, we extracted retrospective EMR data from the respiratory outpatient department of The Children’s Hospital of Zhejiang University School of Medicine. Filtering the data by ICD-10 disease diagnosis code, we retrieved 181,229 medical records of 107,840 patients who were diagnosed with chronic cough in the hospital from August 2019 to November 2020. A total of 2,936 cases didn’t meet the requirements of our training task due to a lack of information and were excluded. The remaining 178,293 records were divided into a training set and a testing set at a ratio of 7:3, with 133,719 records in the model training set and 44,574 in the testing set. We also extracted 5% (6,686 records) of the 133,719 records in the model training set as the validation set for parameter tuning.
Algorithm
The golden standard for the diagnosis of respiratory diseases caused by chronic cough is based on imaging and blood gas test results. However, for the pre-diagnosis stage, since the doctor only conducts inquiry and physical examination, there is no golden standard for prediagnosis. Senior physician can make accurate preliminary disease prediction by considering the details in the chief complaint, such as sputum and nasal mucus, cough sound, fever condition, panting, lung auscultation, etc. Thus we leverage the diagnostic experience of advanced children’s hospitals to provide prediagnosis assist for low-level hospitals.
According to the problem definition, disease prediction is essentially a text classification problem. Since examination data are not available in the prediagnosis stage, the only information available for prediction is that gained from doctor-patient interviews, which can be acquired from EMR text. We designed a system to predict disease type using 7 types of information: age, the patient’s chief complaint, present disease history, past history, family history, history of allergy, and medication history. Considering the excellent performance of the TextCNN (19) model in text classification, we used this model for disease prediction in the prediagnosis stage.
We first input the EMR records into the pretrained language model to obtain embedding vectors to be used as the input of the convolutional layer of our disease prediction model. A 100-dimensional lookup table was then generated in the convolutional layer, which can be denoted as:
Here denotes the cascading operator, and denotes a sequence of words . The convolution operation involves a filter that is applied to a window containing h words to generate features. Feature ci is generated from the window of word Xi to word :
Here f is the activation function, W is the weight of the sequence, and b is the offset. The convolution kernel is then applied to generate feature map for the sentence , and the maximum value of feature map c is taken as the feature of that particular filter. Each kernel has 128 filters, which forms an eigenmatrix of 1*128.
In the convolution stage (Figure 2), the parameters of the pretrained language model were transferred to the disease prediction model through fine-tuning. We set the kernel size as [3, 4, 5] for the disease prediction model. To avoid information loss, the width of the convolution kernel was set as the same as the dimension of the word vector, and each kernel had 128 output channels.
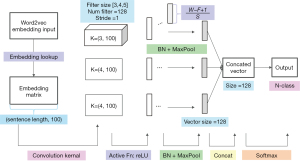
Since the sentences were vectorized by word embedding, we could carry out a convolution scan at character level, which needs to contain all the feature vectors of the characters. Therefore, the width of the convolution kernel should be consistent with the width of embedding vector. Each convolution kernel had 128 filters, and the stride was set to [1, 1]. The rectfier linear unit (ReLU) function was used as the activation function. For network initialization, the Glorot normal distribution initialization method was used. To adjust data distribution, we also added batch normalization (BN) to the network (20). 1-max pooling was used to improve the stability of model. We generated three 128-length feature vectors and concatenated them into a 384-dimensional vector before dropout. Finally, we used a fully connected structure and softmax as the output layer. The formula of the softmax function was as follows:
Here VX denotes the features formed by feature tiling after convolution and pooling, and y denotes the output of disease prediction. i and j denote a certain disease type in the disease set.
Cross entropy was adopted as the loss function of the disease prediction model:
Here tki is the probability that sample k belongs to category i, and yki is the probability that sample k is predicted as category i. m is the size of disease set, and R is the size of the EMR training set.
Model optimization
To tackle the problem of uneven distribution of the training data among different categories of diseases, we used focal loss (21) instead of cross entropy loss for the disease prediction task. The focal loss function improved the accuracy of the model by introducing the parameter and the modulation coefficient and adjusting the shared weight of positive and negative samples while controlling the weight of difficult and easy samples. The loss function of focal loss is represented as follows:
Here represents the probability of model prediction in the classification task:
Statistical analysis
We used precision, recall, and F1-score to measure the performance of algorithms for each disease type in the target category (Table 1). As disease prediagnosis is a multiclassification task, evaluation indexes were calculated separately for each category. The evaluation measurements used for the prediagnosis disease prediction and examination recommendation tasks were both based on a confusion matrix (Table 2). The formula definition of precision, recall, and F1-score was as follows:
Table 1
| Class label | Class name | Records | Disease names | ICD-10 code |
|---|---|---|---|---|
| 0 | AURI | 57318 | Acute upper respiratory infection | J06.900 |
| Upper respiratory infection | J06.900x003 | |||
| 1 | Bronchitis | 38502 | Acute bronchitis | J20.900 |
| Bronchitis | J40.x00 | |||
| Asthmatic bronchitis | J45.901 | |||
| Chronic asthmatic bronchitis | J44.804 | |||
| 2 | Asthma | 15002 | Noncritical bronchial asthma | J45.903 |
| Asthma | J45.900 | |||
| Bronchial asthma | J45.900x001 | |||
| Cough variant asthma | J45.005 | |||
| 3 | Pharyngitis | 12374 | Acute pharyngitis | J02.900 |
| Acute nasopharyngitis | J00.x00 | |||
| Herpangina | B08.501 | |||
| Infective nasopharyngitis | J00.x00x007 | |||
| Pharyngitis | J02.900x004 | |||
| 4 | Pneumonia | 4465 | Bronchopneumonia | J18.000 |
| Acute bronchopneumonia | J18.000a | |||
| Pneumonia | J18.900 | |||
| Acute pneumonia | J18.900a | |||
| 5 | Rhinitis | 4285 | Anaphylactic rhinitis | J30.400 |
| Acute rhinitis | J00.x00 | |||
| Allergic Rhinitis with Asthma | J45.004 | |||
| Allergic rhinitis | J30.400 | |||
| Rhinitis | J31.000x001 | |||
| 6 | Tonsillitis | 2992 | Acute tonsillitis | J03.900 |
| Acute suppurative tonsillitis | J03.901 | |||
| 7 | Laryngitis | 2850 | Acute laryngitis | J04.000 |
| Acute laryngotracheitis | J04.200 | |||
| 8 | Nasosinusitis | 2490 | Nasosinusitis | J32.900, J32.900x001 |
| Acute nasosinusitis | J01.900 | |||
| Chronic nasosinusitis | J32.900 | |||
| 9 | FLU | 2430 | Influenza | J11.101 |
| 10 | FBAO | 2327 | Upper airway cough syndrome | R05.x00 |
| Acute tracheitis | J04.100 | |||
| 11 | Others | 1795 | – | – |
Disease abbreviation: AURI, acute upper respiratory infection; FLU, influenza; FBAO, foreign body airway obstruction.
Table 2
| Category | True value =1 | True value =0 |
|---|---|---|
| Prediction value =1 | TP | FP |
| Prediction value =0 | FN | TN |
Value of the confusion matrix elements: TP, true positive; FP, false positive; FN, false negative; TN, true negative.
For each type of disease in the target category, we respectively calculated the precision, recall, and F1-score metrics for our method and the baseline methods. We then calculated AC, macro average (MA), and weighted average (WA) of precision, recall and F1-score to evaluate the overall effect of the algorithm on the test dataset. The formulas of AC, MA, and WA were as below:
Here n denotes the number of disease categories, Si denotes the score (precision/recall/F1-score) of each category, and Wi denotes the weight of each category, which was calculated by dividing the number of instances of each category Csup port by the total number of test sets CTotal.
Experiment setup
We counted common diseases with cough as the chief complaint in children and reclassified the diseases according to similarity of symptoms as suggested by respiratory experts in our hospital. After incorporating similar subcategories, 12 disease types were finally included as the prediction target categories (Table 1).
For data processing, we extracted the patient’s medical records, including age, chief complaint, present disease history, past history, family history, history of allergy, and medication history. After removing stop words, we concatenated different attributes into a combined text snippet as the patient’s context snippet. Our model then learned through these patient context snippets from the training set.
The entire pipeline was built using a bag of words model in Python version 3.6.2 (Python Software Foundation, Wilmington, DE, USA) with the gensim 3.8.3 package. Machine learning (ML) classifiers and model evaluation were implemented using the scikit-learn version 0.23.2 package (22). Tensorflow version 2.1.0 was used to construct the TextCNN architecture (23).
Examination recommendation model
Dataset
For examination recommendation, we also extracted 181,229 retrospective medical records from the EMR data as the disease prediction dataset. We counted all the examination items for the different disease types. The items with too few cases were filtered out, and those with a total number of more than 500 cases were selected and ranked as the label column. Finally, we choose the top 10 most frequent examination items in pediatric respiratory clinics as the recommended items. These included blood routine examination (blood RT), abnormal lymphocyte detection (AL), hypersensitive C-reactive protein (hs-CRP), CAP allergen test (CAP), immunoglobulin (IGG), complement (alexin), chest normal position piece (chest PA), renal function, stool, and respiratory virus detection. We excluded EMR records without any examinations in the 10 categories and formed a dataset of 42,967 cases. Then, we divided the training set and the test set at a ratio of 3:1 (3 for the model training set and 1 for the testing set). Among the 32,225 cases in the model training set, we extracted 5% (1,611 cases) as the validation set for parameter tuning.
Algorithm
Examination recommendation is essentially a multilabel classification problem. Our model recommends examination items according to the clinical information of patients, such as age, chief complaint, history of present disease, past history, history of allergy, and the diagnosis given by doctors. For training, we concatenate the information in diagnosis and in clinical notes. The model also learns through a TextCNN framework to generate examination recommendation results. Our prediagnosis system can achieve two tasks: first generating the disease prediction result from the disease prediction model; then the predicted disease with the highest probability is taken as part of the input for the examination recommendation model; finally the system will generate examination recommendation results based on the examination recommendation model.
The TextCNN model was also used in our recommendation model, but with binary cross entropy loss as the loss function, since examination recommendation becomes a binary classification problem for every given examination item. The predicted average probability error of each examination category was taken as the overall error of the model, and the parameters were updated using the back propagation algorithm. The formation of our loss function is represented as follows:
Here y is the ground truth status that a certain examination is taken or not taken (1 represents taken, 0 otherwise) of the sample, and denotes the probability that the model predicts the sample as a positive case.
Statistical analysis
We selected 10 common examination items in pediatric chronic cough as the recommended items, and the prediction of each category was binary with a value 0 or 1 (0 meaning that the examination was not recommended, and 1 meaning that it was recommended). For evaluation, we calculated precision, recall, and F1-score for the 0 and 1category (examination undertaken or not undertaken) respectively by integrating the examination recommendation results of different examine items under 0 and 1categories. The calculation method of the precision, recall and F1-score metrics was similar to that of disease prediction. We calculated AC, MA precision, WA precision, MA recall, and WA recall to assess the performance of our examination recommendation method. The formula of AC was different from that of disease prediction:
We designed this metric specially for the examination recommendation task. Here n denotes the number of samples in the EMR dataset. F (n) calculates the prediction AC of each examination category through matching between the predicted result vector and the ground-truth vector. In F (n), the element-wise product of 2 vectors H (Xi) and Xi is calculated. H (Xi) represents the model output vector for sample Xi, while Yi represents the ground-truth examination vector of each sample. Both are one-dimensional arrays whose element is a binary value to show whether the examination is recommended (1 for true and 0 for false). In addition, we also calculated the receiver operating characteristic (ROC) and the area under the curve (AUC) to evaluate the sensitivity and specificity of the algorithm.
Ethical statement
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Academic Ethics Committee of Children's Hospital Affiliated to Zhejiang University School of Medicine (No. 2020-RIB-058). Individual consent for this retrospective analysis was waived.
Results
Disease prediction results
Comparison with state-of-the-art methods
Baseline methods
We compared our method with 4 state-of-the-art baselines that were suitable for text classification on our dataset, including the logistic regression (LR) algorithm (24), the gradient-boosted decision tree (GBDT) algorithm (24), the hierarchical attention networks (HAN) model (25), and the bidirectional encoder representations from transformers (BERT) model (26). The former two are ML methods, while the latter two are DL algorithms widely adopted in NLP tasks. For our method, we used our MSCNN framework, which contained a preliminary embedding process and a TextCNN-based disease prediction process. Training data were divided into 5 folds for cross-validation, and the results are shown in the figures below.
Top-1 result comparison
The output of the prediagnosis disease prediction task was a 12-dimensional vector. Each dimension represented the probability of a certain disease occurring. The component with the maximum value was the top-1 result. Figure 3 shows the AC, recall, and F1-score of top-1 results predicted by the 5 algorithms for the 12 disease categories.
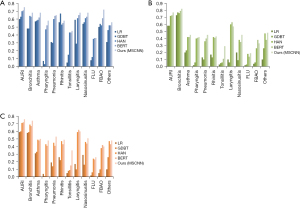
Results of ML methods
The model performance of the 5 algorithms was basically proportional to their feature extraction ability. LR and GBDT ML models showed weak prediction ability. The highest precision values of LR and GBDT for the 12 disease categories were 0.6 and 0.63, respectively, both occurring in the acute upper respiratory infection (AURI) class. The highest recall values of LR and GDBT were 0.73 and 0.77, respectively, both occurring in the bronchitis class. However, for some disease categories, such as pharyngitis, tonsillitis, and FLU, the precision, recall, and F1-score of the LR and GBDT algorithms was below 0.1.
Results of DL methods
Judging from the top-1 result, the prediction effect of the DL methods was obviously better than that of the ML methods. Compared with HAN and BERT, our method (MSCNN) had the strongest prediction ability in this task in terms of the performance on precision, recall, and F1-score metrics. BERT generally performed better than HAN in all categories. The highest precision values of HAN, BERT, and our method in the 12 disease categories were 0.7, 0.71, and 0.75, respectively, all occurring in the AURI class. Our method performed approximately 5% to 10% better than the other 2 DL algorithms on precision. For recall, the highest values of HAN, BERT, and our method were 0.76, 0.79, and 0.82, respectively, all occurring in the bronchitis category. Our method performed approximately 3% to 11% better than the other 2 DL algorithms on recall. For F1-score, the highest values of HAN, BERT, and our method were 0.71, 0.72, and 0.76, respectively, all occurring in the AURI class. The best-performing disease classes for the DL methods appeared to be similar to that for the ML approaches, possibly due to differences in data quality for the different disease categories. In addition, for categories whose data volumes were small [e.g., tonsillitis, FLU, and foreign body airway obstruction (FBAO)], the performance of all DL methods was low.
Comparison of methods with and without data enhancement
In the prediagnosis stage, patients have not yet undergone any examinations, so the information available for disease prediction consists of the patient’s chief complaint, age, and other textual information in EMR notes. The precision value for the top-1 result in our method was 0.6–0.8 in most disease categories. Thus, we added a data enhancement process through the embedding replace method to promote performance. We calculated the term frequency-inverse document frequency (TF-IDF) features of words, and the 5 words with the largest TF-IDF feature values were sorted out as salient words (27). For salient words, we used a pretrained word2vec model to calculate the similarity with words in the lookup table of word2vec and replaced the original words with the words with the highest similarity to generate new enhanced samples.
Table 3 compares the AC, MA precision, WA precision, MA recall, WA recall, MA F1-score, and WA F1-score for the 5 algorithms with and without data enhancement. Since we needed to measure the performance of different methods at the dataset level, we took the MA and WA values of multiple disease classes on precision, recall, and F1-score for the different methods. After data enhancement, the performance of the 4 algorithms was slightly improved, excepting that of the LR algorithm. This may be because the LR algorithm is prone to underfitting, and the enhanced data lead to more serious underfitting problems. However, in general, data enhancement is able to improve performance.
Table 3
| Methods | AC | MA precision | WA precision | MA recall | WA recall | MA F1-score | WA F1-score |
|---|---|---|---|---|---|---|---|
| LR | 0.52 | 0.35 | 0.51 | 0.18 | 0.52 | 0.21 | 0.48 |
| LR + data enhancement | 0.47 | 0.34 | 0.48 | 0.31 | 0.47 | 0.31 | 0.47 |
| GBDT | 0.54 | 0.38 | 0.53 | 0.18 | 0.54 | 0.21 | 0.5 |
| GBDT + data enhancement | 0.54 | 0.44 | 0.54 | 0.32 | 0.53 | 0.34 | 0.53 |
| HAN | 0.62 | 0.51 | 0.61 | 0.42 | 0.62 | 0.45 | 0.61 |
| HAN + data enhancement | 0.63 | 0.54 | 0.62 | 0.42 | 0.63 | 0.46 | 0.62 |
| BERT | 0.64 | 0.54 | 0.61 | 0.44 | 0.65 | 0.48 | 0.65 |
| BERT + data enhancement | 0.64 | 0.55 | 0.62 | 0.44 | 0.67 | 0.47 | 0.65 |
| Ours (MSCNN) | 0.68 | 0.56 | 0.67 | 0.45 | 0.68 | 0.5 | 0.67 |
| Ours (MSCNN) + data enhancement | 0.68 | 0.59 | 0.67 | 0.49 | 0.68 | 0.51 | 0.67 |
Metrics included AC, MA precision, WA precision, MA recall, WA recall, MA F1-score, and WA F1-score. AC, accuracy; MA, macro average; WA, weighted average. Methods include: LR, logistic regression; GBDT, gradient-boosted decision tree; HAN, hierarchical attention networks; BERT, bidirectional encoder representations from transformers; MSCNN, medical-semantic-aware convolution neural network.
Table 4 presents the top-3 AC results (the 3 highest-scored components) of different methods. It also shows clearly that the data enhancement can help to promote prediction AC for both ML and DL methods.
Table 4
| Methods | AC |
|---|---|
| LR | 0.759 |
| LR + Data Enhancement | 0.822 |
| GBDT | 0.789 |
| GBDT + Data Enhancement | 0.821 |
| HAN | 0.907 |
| HAN + Data Enhancement | 0.911 |
| BERT | 0.901 |
| BERT + Data Enhancement | 0.905 |
| Ours (MSCNN) | 0.923 |
| Ours (MSCNN) +Data Enhancement | 0.926 |
Methods include: LR, logistic regression; GBDT, gradient-boosted decision tree; HAN, hierarchical attention networks; BERT, bidirectional encoder representations from transformers; MSCNN, medical-semantic-aware convolution neural network; AC, accuracy.
Ablation study for the pretrained medical language model
To demonstrate the role of the pretrained language model in the whole framework, we conducted an ablation experiment. We compared the performance changes in precision, recall, and F1-score of disease prediction tasks by using the TextCNN model before and after the pretrained medical language model was adopted. As can be seen in Table 5, when we didn’t use the medical-semantic-aware pretrained language model to generate embedding vectors as input for the TextCNN model, the performance of the framework was lower than that of the MSCNN framework under 3 indicators (precision/recall/F1-score) in the 12 disease categories. In most disease categories, the performance improvement achieved using the MSCNN framework was around 20%. The reason for the improvement was that the pretrained language model was trained with medical literature resources; therefore, it had a good semantic awareness of medical-related textual content and was able to generate more accurate semantic representations for the EMR text.
Table 5
| Disease | Precision | Recall | F1-score | |||||
|---|---|---|---|---|---|---|---|---|
| TextCNN | MSCNN | TextCNN | MSCNN | TextCNN | MSCNN | |||
| AURI | 0.56 | 0.75 | 0.59 | 0.77 | 0.58 | 0.76 | ||
| Bronchitis | 0.49 | 0.68 | 0.68 | 0.82 | 0.55 | 0.74 | ||
| Asthma | 0.48 | 0.68 | 0.26 | 0.45 | 0.32 | 0.5 | ||
| Pharyngitis | 0.41 | 0.58 | 0.25 | 0.42 | 0.29 | 0.48 | ||
| Pneumonia | 0.46 | 0.65 | 0.23 | 0.45 | 0.26 | 0.53 | ||
| Rhinitis | 0.37 | 0.58 | 0.23 | 0.43 | 0.33 | 0.49 | ||
| Tonsillitis | 0.27 | 0.45 | 0.11 | 0.29 | 0.17 | 0.35 | ||
| Laryngitis | 0.50 | 0.68 | 0.38 | 0.57 | 0.38 | 0.6 | ||
| Nasosinusitis | 0.49 | 0.68 | 0.28 | 0.45 | 0.34 | 0.51 | ||
| FLU | 0.19 | 0.36 | 0.05 | 0.19 | 0.07 | 0.26 | ||
| FBAO | 0.53 | 0.72 | 0.16 | 0.33 | 0.27 | 0.4 | ||
| Others | 0.39 | 0.56 | 0.22 | 0.48 | 0.30 | 0.48 | ||
Metrics included precision, recall, and F1-score. TextCNN, text convolutional neural network; MSCNN, medical-semantic-aware convolution neural network; AURI, acute upper respiratory infection; FLU, influenza; FBAO, foreign body airway obstruction.
Examination recommendation results
Results of the dataset
Table 6 shows the results of the MSCNN examination recommendation approach for the 10 selected examination categories. As examination recommendation also used the TextCNN model, we did not compare its performance with other algorithms. For each examination item, we first calculated the precision, recall, and F1-score values for positive and negative cases. That is, when a given examination item was undertaken (positive, status =1) or not undertaken (negative, status =0) in the ground-truth data, we calculated its precision, recall, and F1-score. Then, for each examination item, we integrated the positive and negative measures with the MA and WA values on the 3 indicators. The calculation of the indicator AC was demonstrated in the Evaluation Protocol section above.
Table 6
| Examination | AC | MA precision | WA precision | MA recall | WA recall | MA F1-score | WA F1-score |
|---|---|---|---|---|---|---|---|
| Blood RT | 0.88 | 0.84 | 0.87 | 0.55 | 0.88 | 0.56 | 0.83 |
| AL | 0.89 | 0.79 | 0.88 | 0.64 | 0.89 | 0.67 | 0.88 |
| hs-CRP | 0.87 | 0.84 | 0.87 | 0.56 | 0.87 | 0.57 | 0.83 |
| CAP | 0.98 | 0.7 | 0.97 | 0.62 | 0.98 | 0.65 | 0.97 |
| IGG | 0.98 | 0.72 | 0.97 | 0.62 | 0.98 | 0.65 | 0.97 |
| Alexin | 0.98 | 0.72 | 0.97 | 0.62 | 0.98 | 0.65 | 0.97 |
| chest PA | 0.63 | 0.7 | 0.78 | 0.71 | 0.63 | 0.63 | 0.64 |
| Renal function | 0.98 | 0.87 | 0.98 | 0.8 | 0.98 | 0.83 | 0.98 |
| Stool | 0.99 | 0.91 | 0.99 | 0.59 | 0.99 | 0.65 | 0.99 |
| Respiratory virus detection | 0.89 | 0.54 | 0.96 | 0.65 | 0.89 | 0.55 | 0.92 |
Metrics included AC, MA precision, WA precision, MA recall, WA recall, MA F1-score, and WA F1-score. MSCNN, medical-semantic-aware convolution neural network; AC, accuracy; MA, macro average; WA, weighted average; blood RT, routine examination; AL, abnormal lymphocyte detection; hs-CRP, hypersensitive C-reactive protein; CAP, CAP allergen test; IGG, immunoglobulin; chest PA, chest posteroanterior.
In Table 6, we can see that the examination items with the highest AC were not normal examination items like blood RT (AC =0.88) and chest PA (AC =0.63) but uncommon examination items like stool (AC =0.99), IGG (AC =0.98), renal function (AC =0.98) and alexin (AC =0.98). The effectiveness of our inspection recommendation method is reflected in the following two points. (I) Routine examinations like blood RT and chest PA, which doctors often overuse in the treatment of respiratory diseases, do not gain the highest score in our algorithm. Therefore, our method does not recommend frequent items for every patient. (II) For items infrequently used by doctors, our algorithm achieved a very high AC (e.g., stool and alexin). This indicates the effectiveness of our algorithm in helping doctors determine examination plans, as our approach is able to accurately predict the use of infrequent examination items.
Table 7 shows the AC of the entire dataset, as well as the precision, recall, and F1-score values for the 0 and 1 statuses of our prediction results. The total prediction AC for the entire dataset was 93%. The performance was better for negative status than for positive status; thus, our approach is also able to exclude inappropriate examination items.
Table 7
| Status | Precision | Recall | F1-score |
|---|---|---|---|
| 0 | 0.98 | 0.94 | 0.96 |
| 1 | 0.74 | 0.89 | 0.81 |
| AC | 0.93 | ||
| MA | 0.86 | 0.91 | 0.88 |
| WA | 0.94 | 0.93 | 0.93 |
We conducted precision, recall, and F1-score measurements in positive (status =1, examination undertaken) and negative (status =0, examination not undertaken) cases. AC was calculated for the overall dataset. MA precision/recall/F1-score and WA precision/recall/F1-score were calculated through weighted and macro averages of the 10 examination categories. AC, accuracy; MA, macro average; WA, weighted average.
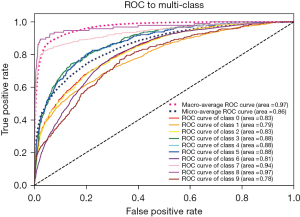
Figure 4 shows the ROC and AUC curves for the 10 examination categories. The prediction performance of the algorithm in the different examination categories was generally even, except that the stool and renal function examination categories were much better. The examination class with the lowest ROC value was chest PA (AUC =0.81). The high performance on AUC reflects the strong sorting ability of our model. The model also showed good ability in the division of positive and negative categories.
Discussion
Related work
As DL methods have developed, more research has investigated intelligent diagnosis methods for respiratory diseases such as pneumonia, COVID-19, and lung cancer. Most of the work in this field has focused on diagnosis in the formal diagnosis stage when medical inspections have been performed. For example, Umar Ibrahim et al. (4) proposed a bidirectional adversarial network-based framework to develop models predictive of COVID-19 versus non-COVID-19 viral pneumonia using CT images. Wang et al. (5) developed a fully automated DL pipeline for the visualization of lesions and the diagnosis of pneumonia caused by COVID-19 on CXR images that was able to discriminate between viral pneumonia caused by COVID-19 and other types of pneumonia. Another study proposed a fully automated DL system (6) for COVID-19 diagnostic and prognostic analysis by routinely used CT images. Yu et al. proposed a DL framework CGNet (28) for classifying CXR images into normal and pneumonia categories. Their model includes feature extraction, graph-based feature reconstruction, and classification. All the above works relied on image data for learning.
To the best of our knowledge, research on intelligent diagnosis assistance in the prediagnosis stage has not yet been undertaken, and this is the first study to investigate intelligent prediagnosis for pediatric chronic cough. Wagner et al. (29) developed an intelligent data enhancing platform for the diagnosis of COVID-19. The platform applied data enhancement algorithms and deep neural networks (DNNs) to analyze the comparison of disease symptoms between COVID-19 patients and healthy patients in EMR records in the week prior to polymerase chain reaction (PCR) testing to discover derivative features of COVID-19 in early stage of the disease. Kam et al. (12) extracted EMR data of multiple biological signal variables from the MIMIC II database and built a prediction network using a DNN model to facilitate the early detection of sepsis. Since these variables were updated every few hours, the authors added a long short-term memory (LSTM) neural network for the learning of time-series data. Kam et al. (12) also used EMR data combined with time-series data and a recurrent neural network (RNN) to predict Parkinson’s disease. Using RNN architecture, their algorithm encoded the similarity between the patterns of 2 sequences in patient treatment records to predict disease risk. Mullenbach et al. (30) proposed an approach using a CNN to automatically assign specific ICD-9 codes to discharge summaries of intensive care unit (ICU) inpatients. The authors used a per-tag attention mechanism to learn document representations for each tag. Their convolution attention multiple label (CAML) model interpreted the classification of diseases from clinical records to produce diagnostic codes and obtained good results on the MIMIC-II and MIMIC-III datasets. Wang et al. (14) constructed a prediction model using EHRs and a knowledge-based CNN to estimate the distant recurrence probability of patients with breast cancer. By mapping the documents to a concept space, they generated embeddings with concept unified identifiers (CUI) tags and transmitted the embeddings to a knowledge-guided CNN (K-CNN) for prediction. Qiu et al. (31) applied DNN and CNN networks to free text pathological reports to automatically analyze the primary cancer and lateral positions of breast cancer and lung cancer. Irvin et al. (32) developed an automatic labeling NLP tool that was able to identify radiological reports of 14 different varieties of thoracic diseases with greater accuracy than human annotators. Although there are many diagnosis assistance studies using EMR data, there is still no research leveraging doctor-patient interviews and EMR data for diagnosis assistance in the prediagnosis stage.
In this paper, we conducted research on intelligent prediagnosis for pediatric chronic cough using textual EMR data and an NLP convolution neural network. CNN algorithms have been successfully applied to a variety of NLP tasks, such as text classification (33), sentiment analysis (34), and language modeling (35). Other recent work has combined convolution and attention mechanisms (36,37). Our work differs to these studies in that we take the specialty of medical data into account. We combined convolution with a language model that was pretrained on a medical literature corpus for more accurate semantic representation of medical-related content. This model could then be rapidly transferred to different downstream AI tasks for learning.
Discussion of approach
We compared our method with two ML methods (LR and GBDT) and two DL methods (BERT and HAN). The ML models showed weak prediction ability compared to the DL methods. The reasons for this may arise from three factors. First, ML algorithms are prone to deviation due to unbalanced sample distribution. In our dataset, the number of cases in the different disease categories was not even. AURI and bronchitis had more than 10,000 cases, while the case numbers of 4 categories (asthma, pharyngitis, pneumonia, and rhinitis) were between 300 and 1,000 cases, and those of 5 categories (tonsillitis, laryngitis, lasosinusitis, FLU, and FBAO) were lower than 1,000. In addition, the top-1 AC result of the intelligent prediagnosis task was strongly related to the number of the training cases. Second, ML methods do not learn features deeply enough and often encounter overfitting and underfitting problems. This is a common problem with ML approaches. Third, data quality unevenness also contributed to the differences in the results. Since our learning methods were based on information from real historical EMR records, the information in the EMR text was not always of a high quality. In some cases, the description of the chief complaint was very simple, with much information missing. Therefore, the quality of the textual data was uneven, and this lead to the poor performance of the ML algorithms in text classification.
BERT and HAN models are common DL models in text classification prediction. Generally speaking, the BERT model performs better, since the HAN model is not able to find minority groups in the data, and its performance is biased by the majority class. In our task, the performance of BERT was inferior to our method, which may be attributed to two causes. First, BERT has too many parameters and is more suitable for a refined classification task with more detailed data content. TextCNN performs better with coarse-grained classification problems and is thus more suitable for EMR text, which usually lacks detail. Second, BERT does not use embedding vectors that are aware of medical content. In contrast, our MSCNN framework used a pretrained language model to generate medical-semantic-aware representations, which greatly improved classification. This was confirmed in the subsequent ablation experiments.
To promote diagnostic accuracy, we also added a pretrained medical language model and a data enhancement process before disease prediction. We used a medical literature corpus to train a model that could generate word embeddings for input text. The ablation studies of the pretrained medical language model showed an approximately 20% promotion in performance, as the awareness of medical semantic content was elevated through the pretrained medical language model. For data enhancement, we calculated TF-IDF features of words and generated salient words as input. However, this did not lead to an obvious improvement in performance.
Limitations of our method
This work had several limitations. First, the approach applied embedding of concatenated EMR text as input to the disease prediction model by relying on the pretrained medical language model. One limitation is that we might carry along modeling errors that exist in the pretrained language model. Another limitation of the disease prediction model is that it does not use attention mechanisms for medical entities. In the future, to improve the prediction AC, we could leverage an external knowledge database to realize the medical-related entities and add attention mechanism for those terms. Another issue is related to the data quality. In EMR records, the chief complaint information is often incomplete, which obviously affects the prediction AC. Including information from the physical examination performed by doctors in the prediagnosis stage may improve the effect of the intelligent prediagnosis model. Another limitation is that the proposed MSCNN framework is restricted to solving prediction problems using textual data.
Conclusions
We constructed an intelligent prediagnosis system for chronic cough in children to facilitate disease prediction and examination recommendation using clinical information in EMR records. We proposed an MSCNN framework for the overall system. We first trained a medical language model using a medical literature corpus to generate more accurate semantic representations for downstream AI tasks. This language model benefitted succeeding tasks through transfer learning. We then built a neural network based on the TextCNN model to achieve disease prediction and examination recommendation. The ablation study showed that a pretrained language model plays a large role in promoting the semantic understanding of medical content in EMR data. Our approach showed good results on real clinical datasets both in disease prediction and examination recommendation tasks. For disease prediction, our approach outperformed the 4 baseline methods (including two ML methods and two DL methods) on all the metrics. For examination recommendation, our MSCNN framework also achieved high AC in recommending examination items. However, for datasets with small volume, the precision and recall performance of our method was not as good. Improving the prediction AC for small-volume datasets will be the focus of future research. More sophisticated attention mechanisms and deeper expertise knowledge could be added to our neural network in the future. Further development of the model with advanced NLP and DL techniques will enable the model to achieve a more accurate performance. Further investigation is needed to validate the clinical application of our model.
Acknowledgments
Funding: This work was supported by the National Key R&D Program of China (No. 2019YFE0126200) and the National Natural Science Foundation of China (No. 62076218). The prediagnosis system for pediatric chronic cough proposed in this work is part of the design of the technical framework of the funding. However, the writing is irrelevant to the funding body.
Footnote
Reporting Checklist: The authors have completed the STARD reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-22-275/rc
Data Sharing Statement: Available at https://tp.amegroups.com/article/view/10.21037/tp-22-275/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-22-275/coif). Hongjian Zhang, Siyu Chen, Qianhui Zhong, and Yulan Xie report that they are from Avain (Hangzhou) Technology Co., LTD.; Dejian Wang is from Hangzhou Healink Technology. The other authors declare that they have no conflicts of interest.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Academic Ethics Committee of Children’s Hospital Affiliated to Zhejiang University School of Medicine (No. 2020-RIB-058). Individual consent for this retrospective analysis was waived.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chang AB, Berkowitz RG. Cough in the pediatric population. Otolaryngol Clin North Am 2010;43:181-98. xii. [Crossref] [PubMed]
- Morice AH, Millqvist E, Bieksiene K, et al. ERS guidelines on the diagnosis and treatment of chronic cough in adults and children. Eur Respir J 2020;55:1901136. [Crossref] [PubMed]
- Weinberger M, Hurvitz M. Diagnosis and management of chronic cough: similarities and differences between children and adults. F1000Res 2020;9:eF1000 Faculty Rev-757.
- Umar Ibrahim A, Ozsoz M, Serte S, et al. Convolutional neural network for diagnosis of viral pneumonia and COVID-19 alike diseases. Expert Syst 2021; Epub ahead of print. [Crossref] [PubMed]
- Wang G, Liu X, Shen J, et al. A deep-learning pipeline for the diagnosis and discrimination of viral, non-viral and COVID-19 pneumonia from chest X-ray images. Nat Biomed Eng 2021;5:509-21. [Crossref] [PubMed]
- Fernandes V, Junior GB, de Paiva AC, et al. Bayesian convolutional neural network estimation for pediatric pneumonia detection and diagnosis. Comput Methods Programs Biomed 2021;208:106259. [Crossref] [PubMed]
- Mahin M, Tonmoy S, Islam R, et al. Classification of COVID-19 and Pneumonia Using Deep Transfer Learning. J Healthc Eng 2021;2021:3514821. [Crossref] [PubMed]
- Ibrahim DM, Elshennawy NM, Sarhan AM. Deep-chest: Multi-classification deep learning model for diagnosing COVID-19, pneumonia, and lung cancer chest diseases. Comput Biol Med 2021;132:104348. [Crossref] [PubMed]
- Duan X, Guo Y, Radiology DO. Application and prospect of health technology assessment in medical imaging. International Journal of Medical Radiology 2017;40:133-6.
- Olgar T, Sahmaran T. Establishment of radiation doses for pediatric X-ray examinations in a large pediatric hospital in Turkey. Radiat Prot Dosimetry 2017;176:302-308. [Crossref] [PubMed]
- Hess CB, Thompson HM, Benedict SH, et al. Exposure Risks Among Children Undergoing Radiation Therapy: Considerations in the Era of Image Guided Radiation Therapy. Int J Radiat Oncol Biol Phys 2016;94:978-92. [Crossref] [PubMed]
- Kam HJ, Kim HY. Learning representations for the early detection of sepsis with deep neural networks. Comput Biol Med 2017;89:248-55. [Crossref] [PubMed]
- Chao C, Cao X, Jian L, et al. An RNN Architecture with Dynamic Temporal Matching for Personalized Predictions of Parkinson's Disease. Proceedings of the 2017 SIAM International Conference on Data Mining, Houston, Texas, USA, April 27-29, 2017. SDM 2017:198-206.
- Wang H, Li Y, Khan SA, et al. Prediction of breast cancer distant recurrence using natural language processing and knowledge-guided convolutional neural network. Artif Intell Med 2020;110:101977. [Crossref] [PubMed]
- Mikolov T, Chen K, Corrado G, et al. Efficient Estimation of Word Representations in Vector Space. 1st International Conference on Learning Representations, Scottsdale, Arizona, USA, May 2-4, 2013. Workshop Track Proceedings. arXiv:1301.3781v3
- Mao Y, Fung KW. Use of word and graph embedding to measure semantic relatedness between Unified Medical Language System concepts. J Am Med Inform Assoc 2020;27:1538-46. [Crossref] [PubMed]
- Goldberg Y, Levy O. word2vec Explained: deriving Mikolov et al.’s negative sampling word-embedding method. CoRR abs/1402.3722 (2014).
- Mikolov T, Sutskever I, Chen K, et al. Distributed Representations of Words and Phrases and their Compositionality. Proceedings of the 2013 Conference on Advances in Neural Information Processing Systems 26, Lake Tahoe, Nevada, United States, December 5-8, 2013. NIPS 2013: 3111-3119.
- Zhang Y, Wallace BC. A Sensitivity Analysis of (and Practitioners' Guide to) Convolutional Neural Networks for Sentence Classification. CoRR abs/1510.03820 (2015).
- Ioffe S, Szegedy C. Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. CoRR abs/1502.03167 (2015).
- Lin TY, Goyal P, Girshick R, et al. Focal Loss for Dense Object Detection. IEEE International Conference on Computer Vision (ICCV) 2017, Venice, Italy, October 22-29, 2017. ICCV 2017: 2999-3007.
- Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research 2011;12:2825-2830.
- Abadi M, Barham P, Chen J, et al. TensorFlow: A system for large-scale machine learning. CoRR abs/1605.08695(2016).
- Bishop CM. Pattern recognition and machine learning. New York: Springer, 2006: 359-418.
- Yang Z, Yang D, Dyer C, et al. Hierarchical Attention Networks for Document Classification. Proceedings of the 2016 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, San Diego California, USA, June 12-17, 2016. HLT-NAACL 2016: 1480-1489
- Vaswani A, Shazeer N, Parmar N, et al. Attention Is All You Need. Proceedings of the 2017 Conference on Advances in Neural Information Processing Systems 30, Long Beach, CA, USA, December 4-9, 2017. NIPS 2017: 5998-6008.
- Jalilifard A, Caridá V, Mansano A, et al. Semantic Sensitive TF-IDF to Determine Word Relevance in Documents. CoRR abs/2001.09896 (2020).
- Yu X, Wang SH, Zhang YD. CGNet: A graph-knowledge embedded convolutional neural network for detection of pneumonia. Inf Process Manag 2021;58:102411. [Crossref] [PubMed]
- Wagner T, Shweta F, Murugadoss K, et al. Augmented curation of clinical notes from a massive EHR system reveals symptoms of impending COVID-19 diagnosis. Elife 2020;9:58227. [Crossref] [PubMed]
- Mullenbach J, Wiegreffe S, Duke J, et al. Explainable Prediction of Medical Codes from Clinical Text. Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long Papers). 2018. NAACL-HLT 2018:1101-1111.
- Qiu JX, Yoon HJ, Fearn PA, et al. Deep Learning for Automated Extraction of Primary Sites From Cancer Pathology Reports. IEEE J Biomed Health Inform 2018;22:244-51. [Crossref] [PubMed]
- Irvin J, Rajpurkar P, Ko M, et al. CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison. The 33rd AAAI Conference on Artificial Intelligence, Honolulu, Hawaii, USA, January 27 - February 1, 2019. AAAI 2019: 590-597.
- Kim Y. Convolutional Neural Networks for Sentence Classification. Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), Doha, Qatar, October 25-29, 2014. EMNLP 2014: 1746-1751.
- Santos C, Gattit M. Deep Convolutional Neural Networks for Sentiment Analysis of Short Texts. Proceedings of the 25th International Conference on Computational Linguistics, Dublin, Ireland, August 23-29, 2014. COLING 2014:69-78.
- Dauphin YN, Fan A, Auli M, et al. Language Modeling with Gated Convolutional Networks. Proceedings of the 34th International Conference on Machine Learning (ICML), Sydney, NSW, Australia, 6-11 August, 2017. ICML 2017: 933-941.
- Tong N, Tang Y, Chen B, et al. Representation learning using Attention Network and CNN for Heterogeneous networks. Expert Systems with Applications 2021;185:115628. [Crossref]
- Ramaswamy SL, Chinnappan J. RecogNet-LSTM+CNN: a hybrid network with attention mechanism for aspect categorization and sentiment classification. Journal of Intelligent Information Systems 2022;58:379-404. [Crossref]


