
A paediatric dysembryoplastic neuroepithelial tumour (DNET) with deregulated stem cell markers: a case report
Introduction
Dysembryoplastic neuroepithelial tumours (DNETs) are rare intracranial tumours that may have inconsistencies in disease progression (1,2). Several recent studies have described the molecular mutations and deregulations of a few DNET cases (3-6). Out of all 283 published DNET cases, 143 were paediatric patients (age: 0–14 years; supplementary table available at: https://cdn.amegroups.cn/static/public/tp-22-19-1.xlsx). Out of 63 DNA-examined paediatric DNET cases, 33 samples were shown to have fibroblast growth factor receptor 1 (FGFR1) variants. Overall, mutations in the FGFR1 gene have been detected in 32 different types of tumours (7). Given the complexity and heterogeneity of most central nervous system tumours (CNSTs), targeting a single gene is unlikely to be highly effective in improving survivorship (8). Thus, we investigated the status of cancer stem cell (CSC) genes associated with resilience and drug resistance in a paediatric DNET. CSCs are cancer cells that utilise stem cell pathways, enhance tumorigenesis and contribute to drug resistance, and their associated markers have been identified in several CNSTs (9-18). Data on the status of CSCs in DNETs have not been published. As such, this report presents data on several deregulated CSC genes and their variants that may be associated with a predisposition for DNET. We present the following article in accordance with the CARE reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-22-19/rc).
Case presentation
All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the patient’s parents or legal guardians for publication of this case report. A copy of the written consent is available for review by the editorial office of this journal. This work was approved by the Ethical Board of King Abdulaziz University Hospital (board registration No. at the National Committee of Bio- and Medical Ethics: HA-02-J-008; project reference No. 976-12).
Patient information and clinical findings
A 6-year-old boy presented with drug-resistant focal epilepsy since the age of 2 years. He is the only child for a non-consanguineous couple and the product of an uneventful pregnancy that was delivered via caesarean section. The child’s first 2 years of life were uneventful, with healthy psychomotor development without a history of febrile illness or trauma and no family history of epilepsy.
The beginning of his seizures was brief absence-like, which lasted for 15–20 s, occurring at a frequency of 4–5 times per week. Subsequently, the seizures became prolonged and of focal onset daily with an impaired conscious level with secondary generalisation occasionally once or twice a month. The patient initially had a good response with valproate for 6 months, but his seizures recurred, and several antiepileptic medications were added, with no positive response. Subsequently, levetiracetam, clobazam and carbamazepine were added, resulting in partial control. However, the patient still suffered several seizures a week. The patient’s physical and neurological examinations were unremarkable.
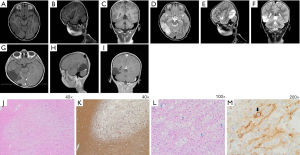
An electroencephalogram (EEG) showed important lateralising interictal epileptiform discharges in the posterior right hemisphere. Long-term video-EEG monitoring revealed that the focal impaired-awareness seizures coincided with ictal EEG activities arising from a right posterior hemispheric lesion. A magnetic resonance imaging (MRI) scan revealed an extensive lesion in the right posterior temporoparietooccipital region (PTR), disclosing hyper T2 signal intensity and hypo T1 signal intensity without contrast enhancement or mass effect. The lesion measured 6.0 cm × 3.0 cm × 2.5 cm, demonstrating focal cortical thickening and a blurred grey-white matter junction (Figure 1A-1F). The preoperative diagnoses were considered low-grade tumour-like DNET or focal cortical dysplasia (FCD). The child underwent right posterior temporoparietooccipital neuronavigation-assisted craniotomy in January 2016. Complete neuronavigation-assisted microsurgical resection was performed and confirmed by postoperative MRI (Figure 1G-1I). A histopathological examination of paraformaldehyde-fixed sections showed pathological features consistent with DNET. The examination revealed a well-demarcated, cortically located multinodular lesion (Figure 1J,1K), forming the so called “specific glioneuronal element”. This element is composed of oligodendrocyte-like cells (OLCs) with round nuclei and clear perinuclear cytoplasmic halos, which are aligned in columns along anuclear zones of neuronal processes containing small blood vessels that are oriented perpendicularly to the cortical surface. The columns are separated by mucin-containing microcysts and a few scattered floating neurons (Figure 1L). Synaptophysin immunohistochemistry staining is positive in the floating neurons and neuropil-like matrix; contrastingly, small OLCs are synaptophysin negative (Figure 1M). The patient remained seizure-free until December 2020.
Molecular and biological analyses
DNET fresh frozen tissue is highly positive for numerous CSC markers
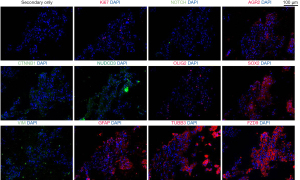
Consecutive sections of the patient’s fresh frozen tumour were cut and immunofluorescence stained to detect CSC markers previously associated with CNSTs (13), as shown in Figure 2, Figure S1 and Supplementary Methods File 1 (Appendix 1). Staining revealed vast differences for Ki67 positivity across tumour regions. Positive cells were grouped into particular regions of the tumour, with 2.4%±0.9% in positive regions. Low expression was detected for Notch receptor 1 (NOTCH1; 13.6%±6.2%) and nuclear SRY-Box transcription factor 2 (SOX2; 6.2%±2.8%). Medium expression was detected for catenin beta 1 (CTNNB1; 34.8%±16.8%), NUDC domain containing 3 (NUDCD3; 42.0%±29.1%), oligodendrocyte transcription factor 2 (OLIG2; 37.9%±7.5%). High expression was detected for anterior gradient 2 (AGR2; 67.2%±18.3%), glial fibrillary acidic protein (GFAP; 95.3%±1.6%), vimentin (VIM; 93.7%±3.6%), tubulin beta 3 class III (TUBB3; 81.7%±9.0%) and the Wnt-signalling-related protein frizzled class receptor 9 (FZD9; 79.0%±14.4%). No expression was detected for nestin (−), prominin 1 (CD133) (−), P53 (−), or SRY-box transcription factor 11 (Sox11) (−) (images not shown).
DNET-retrieved cell line’s resistance to cisplatin and expression of tumorigenic and CSC markers
In order to investigate whether the presence of CSCs in the DNET tissue might have any favourable selection for survival, a DNET cell line was retrieved from the fresh tumour mass and investigated for its drug resistance and CSC marker expression (Figure 3). Cells were culture adapted, had an average doubling time (DT) of 4±0.3 days, a Ki67 expression of 14.8%±5.7%, and were grown to at least passage 15. In order to ensure the cell line was representative of the tumour, the percentage similarity of single nucleotide variants (SNVs) in exomes for DNA collected from the cell line and the corresponding tissue was compared with the percentage similarity of SNVs in exomes for DNA collected from the cell line and non-corresponding tissues (Figure 3B). The previously published cell line, Jed38_MN (12,19), had 98.5% similarity between SNVs in the cell line and its corresponding tissue. It had an average of 56.3% similarity with SNVs in exomes of 13 other meningioma tissues retrieved from other patients. In comparison, the Jed99_DNET cell line had an 89.2% similarity between SNVs in the cell line and its corresponding tissue, a 32.4% average similarity with SNV in the exomes of 14 meningioma tissues retrieved from other patients, and 16.4% average similarity with SNVs detected in the blood of the aforementioned patient. Statistical analysis indicated that the percentage similarity between SNVs in the exomes of DNA collected from the cell line and their corresponding tissues is significantly higher than the percentage similarity of SNVs in the exomes of DNA collected from the cell line and non-corresponding tissues (P<0.001).
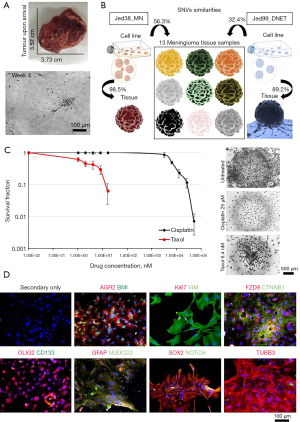
Next, the Jed99_DNET cell line was subjected to the clonogenic assay to check for its ability to form clones in culture and resist drug treatment, both characteristics of aggressiveness. Drug treatment with cisplatin and taxol for passages 8, 10 and 12 indicated an IC50 of 7.1 µM for cisplatin and an IC50 of 3.04 nM for taxol (Figure 3C); notably, several clones survived at doses higher than IC50. In order to address whether the retrieved cells were tumorigenic, cells were immunofluorescence stained for the tumorigenic proto-oncogene (BMI) and metastatic marker (AGR2); results were highly positive for both (Figure 3D). Cells also had high expression (>50%) for several CSC markers, including VIM, FZD9, CTNNB1, GFAP, NUDCD3, TUBB3 and OLIG2 (particularly localised in the nuclear envelope). Notably, the expression of NOTCH1 and nuclear SOX2 were low (<20%), and CD133 was not expressed.
Whole exome sequencing (WES) analysis of the tissue, cell line and blood (TCB)
Having detected deregulated expressions of important CSC markers in both the tissue and corresponding cell line, we investigated the presence of in-common TCB (14) variants for genes listed in the common catalogue of somatic mutations in cancer (COSMIC) database, identified in the SCDevDB database (7,20). The pipeline used for a variant call format (VCF) analysis is shown in Figure 4. Supplementary tables (available at https://cdn.amegroups.cn/static/public/tp-22-19-2.xlsx) and Figure S2 show the COSMIC variants for each sample, their basic features and affected pathways. Supplementary table (available at https://cdn.amegroups.cn/static/public/tp-22-19-3.xlsx) shows selected rare COSMIC variants with damaging coding consequences present in both the tissue and the corresponding cell line Jed99_DNET, but not in the blood (TCnB). This table shows details for somatic related variants including a variant in the insulin growth factor-2 binding protein 3 (IGF2BP3) gene.
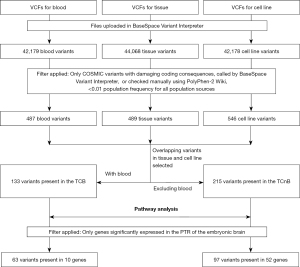
The final filtration selected TCB-COSMIC variants for genes previously published to be relevant for the development of the PTR (20), since the tumour was located in PTR. Ten COSMIC-PTR genes were identified (Table 1); these included the cell division cycle 27 (CDC27) and C-terminal binding protein 2 (CTBP2) genes.
Table 1
| Gene Symbol | Description | Function | COSMIC |
|---|---|---|---|
| ATN1 | Atrophin 1 | BP: GO:0051402 neuron apoptotic process | COSM431781 |
| CDC27 | Cell division cycle 27 | BP: GO: 0045842 positive regulation of mitotic metaphase/anaphase transition; HGS: (M5893) HALLMARK MITOTIC SPINDLE; (M5901) HALLMARK G2M CHECKPOINT; (M5945) HALLMARK HEME METABOLISM | COSM4130311, COSM4130259, COSM5453105, COSM4268063, COSM4130195, COSM4130201, COSM4130207, COSM4130213, COSM4130215, COSM4130217, COSM436751, COSM5611630, COSM4130221, COSM6291465, COSM4130241, COSM4130243, COSM4130245, COSM5764327, COSM3742283, COSM4130257, COSM4130301, COSM4130307, COSM4130309, COSM4130313 |
| CTBP2 | C-terminal binding protein 2 | BP: GO: 0048386 positive regulation of retinoic acid receptor signaling pathway; CP: (M5493) WNT SIGNALING | COSM5620575, COSM5468715, COSM5346315, COSM145279, COSM5021689, COSM3773526, COSM5620563, COSM5620565, COSM213712, COSM328281, COSM2021368, COSM4175021, COSM5703489, COSM5764563, COSM4144437, COSM4144443, COSM1603121, COSM4610155, COSM1600226, COSM4144449, COSM4144451, COSM4144455 |
| FRG1 | FSHD region gene 1 | BP: GO: 0006364 rRNA processing | COSM6029312, COSM4005732 |
| HYDIN | HYDIN axonemal central pair apparatus protein | BP: GO: 1904158 axonemal central apparatus assembly | COSM146271, COSM5958323, COSM1379438, COSM4129291 |
| IGSF3 | Immunoglobulin superfamily member 3 | BP: GO: 0032808 lacrimal gland development; HGS: (M5945) HALLMARK HEME METABOLISM | COSM4142216 |
| KMT2C | Lysine methyltransferase 2C | BP: GO: 0097692 histone H3-K4 monomethylation | COSM216053, COSM4161993, COSM4162005, COSM4162009 |
| NCOR1 | Nuclear receptor corepressor 1 | BP: GO: 0045820 negative regulation of glycolytic process; CP: (M13) PID ERBB4 PATHWAY; (M288) PID HES HEY PATHWAY; (M17) PID NOTCH PATHWAY | COSM5574208 |
| PABPC1 | Poly(A) binding protein cytoplasmic 1 | BP: GO: 2000622 regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay; HGS: (M5949) HALLMARK PEROXISOME; (M5926) HALLMARK MYC TARGETS V1 | COSM6201671, COSM485880, COSM6276973 |
| VWF | von Willebrand factor | BP: GO:0007597 blood coagulation, intrinsic pathway; CP: (M3008) NABA ECM GLYCOPROTEINS; HGS: (M5946) HALLMARK COAGULATION; (M5915) HALLMARK APICAL JUNCTION | COSM6417961 |
COSMIC codes last checked 01/11/2021. All selected genes were significantly associated with the development of the posterior temporoparietooccipital (occipital lobe, parietal lobe, temporal lobe) region of the brain during embryogenesis (P<0.01) (20). All attributed functions were identified by Metascape. BP, biological process (GO); CP, canonical pathways; COSMIC, catalogue of somatic mutations in cancer; DNET, dysembryoplastic neuroepithelial tumour; GO, Gene Ontology; HGS, hallmark gene sets; PTR, posterior temporoparietooccipital region; TCB, tissue, cell line and blood.
Discussion
Our case report elucidated the combined deregulated expression of several CSC markers in a paediatric DNET. Immunofluorescence analysis of the tissue and its corresponding cell line showed positive expressions of several CSC markers. High expressions of GFAP, microtubule-associated protein 2 and synaptophysin were previously observed in reported malignant transformation cases of DNETs (21,22). It is worth mentioning that the positivity of a single marker, such as GFAP, cannot be considered indicative of the differentiation into the glial pathway since also OLIG2 and TUBB3 were positive in the corresponding consecutive sections. Thus, in this case, the triple positivity more likely reflects the progenitor-like state of cancer cells, an observation previously seen in meningioma tissues (12).
The biological characteristics of the corresponding primary cell line, Jed99_DNET, displayed relatively well-established in vitro cell growth. The comparative analysis of the SNVs indicated that DNA collected from the primary cell line is highly representative of the corresponding tissue’s DNA. Critically, cells were highly positive for BMI and AGR2, indicating that the recovered cells were tumorigenic. Additionally, several tested stem cell markers were highly positive and mimicked patterns seen in situ. Consistent with the notion that CSCs are associated with drug resistance, early passage cells were also relatively drug-resistant, particularly to cisplatin.
Ten TCB-COSMIC-PTR genes had damaging variants; the most altered gene, CDC27, possessed 24 variants. CDC27 has an important role as a positive regulator of the mitotic metaphase/anaphase transition and has been shown to be a critical subunit of the anaphase-promoting complex/cyclosome (APC) (23). Two well-studied functional regions of the protein were identified, namely, the anaphase-promoting complex 3 (ApC3) and tetratricopeptide repeat (TPR) regions, which are needed to stabilise interactions with the APC complex (24). The COSMIC database includes several deregulations and mutations of this gene that are associated with embryonal tumours, suggesting a critical and primal role of this gene in driving paediatric tumorigenesis (7,25,26). Furthermore, previously identified mutations in CDC27 were shown to significantly correlate with progressive disease, low overall survival and disease-free survival (23,27). The second gene with the most frequent TCB variants was the CTBP2 gene. The protein is a transcriptional corepressor capable of sensing the levels of cytosolic nicotinamide adenine dinucleotide (NAD) + hydrogen (H) (NADH) and is thought to reside in the nucleus. The deletion of this gene was shown to lead to embryonic lethality, and the involvement of CTBP2 in the Wnt signalling pathway have been shown (28). Further functional large cohort studies are necessary to clarify whether either CDC27 or CTBP2 are critical markers for aggressive DNETs.
Unfortunately, one limitation is the inability to test a large number of samples under the same experimental conditions, as these tumours are rare. In our sample, no COSMIC variations were found in the tissue’s DNA for FGFR1, BRAF or NF1 genes, which is consistent with the aforementioned information mentioned in the introduction and the supplementary table (available at https://cdn.amegroups.cn/static/public/tp-22-19-1.xlsx), as not all histopathologically identified paediatric DNETs have mutations in these genes. However, several variants were detected in other genes that most likely have redundant functions in pathways associated with the aforementioned genes, which may have a parallel effect to FGFR1 mutations. For example, a TCnB COSMIC variant was detected in the IGF2BP3 gene. Both FGFR1 and IGF2BP3 influence the phosphatidylinositol 3-kinase/mitogen-activated protein kinase (PI3K/MAPK) pathway (29,30), therefore perhaps mutations in these genes may have redundant effects on this pathway, which may prove to be important in DNETs.
To our knowledge, the work described in this report assesses for the first time the presence of CSCs in a paediatric DNET. High expressions of several CSC genes were observed in both in situ and the corresponding cell line. Primary cells were relatively drug-resistant, particularly to cisplatin. Using WES analysis, while focusing on the TCB variants of essential developmental genes of the brain’s PTR, identified several genes (e.g., the mitotic regulator gene CDC27 and the Wnt signalling-related gene CTBP2). Further functional large cohort studies are necessary to clarify the diagnostic and prognostic applications of these observations.
Acknowledgments
We would like to thank the KFMRC administration and technical departments for their support, the former director Dr. Ghazi Damanhouri and Dr. Mohammed Jan from the Paediatric Department at KAU Hospital.
Funding: This work was funded by the Dean of Scientific Research, King Abdulaziz University, KSA (G: 489-141-1440).
Footnote
Reporting Checklist: The authors have completed the CARE reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-22-19/rc
Peer Review File: Available at https://tp.amegroups.com/article/view/10.21037/tp-22-19/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-22-19/coif). The authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chiang JCH, Harreld JH, Tanaka R, et al. Septal dysembryoplastic neuroepithelial tumor: a comprehensive clinical, imaging, histopathologic, and molecular analysis. Neuro Oncol 2019;21:800-8. [Crossref] [PubMed]
- Luzzi S, Elia A, Del Maestro M, et al. Dysembryoplastic Neuroepithelial Tumors: What You Need to Know. World Neurosurg 2019;127:255-65. [Crossref] [PubMed]
- Lucas CG, Gupta R, Doo P, et al. Comprehensive analysis of diverse low-grade neuroepithelial tumors with FGFR1 alterations reveals a distinct molecular signature of rosette-forming glioneuronal tumor. Acta Neuropathol Commun 2020;8:151. [Crossref] [PubMed]
- Rivera B, Gayden T, Carrot-Zhang J, et al. Germline and somatic FGFR1 abnormalities in dysembryoplastic neuroepithelial tumors. Acta Neuropathol 2016;131:847-63. [Crossref] [PubMed]
- Surrey LF, Jain P, Zhang B, et al. Genomic Analysis of Dysembryoplastic Neuroepithelial Tumor Spectrum Reveals a Diversity of Molecular Alterations Dysregulating the MAPK and PI3K/mTOR Pathways. J Neuropathol Exp Neurol 2019;78:1100-11. [Crossref] [PubMed]
- Zakrzewska M, Gruszka R, Stawiski K, et al. Expression-based decision tree model reveals distinct microRNA expression pattern in pediatric neuronal and mixed neuronal-glial tumors. BMC Cancer 2019;19:544. [Crossref] [PubMed]
- Tate JG, Bamford S, Jubb HC, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res 2019;47:D941-7. [Crossref] [PubMed]
- Melcher V, Kerl K. The Growing Relevance of Immunoregulation in Pediatric Brain Tumors. Cancers (Basel) 2021;13:5601. [Crossref] [PubMed]
- Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 2014;344:1396-401. [Crossref] [PubMed]
- Venteicher AS, Tirosh I, Hebert C, et al. Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science 2017;355:eaai8478. [Crossref] [PubMed]
- Zhang L, He X, Liu X, et al. Single-Cell Transcriptomics in Medulloblastoma Reveals Tumor-Initiating Progenitors and Oncogenic Cascades during Tumorigenesis and Relapse. Cancer Cell 2019;36:302-318.e7. [Crossref] [PubMed]
- Alamir H, Alomari M, Salwati AAA, et al. In situ characterization of stem cells-like biomarkers in meningiomas. Cancer Cell Int 2018;18:77. [Crossref] [PubMed]
- Khan IN, Ullah N, Hussein D, et al. Current and emerging biomarkers in tumors of the central nervous system: Possible diagnostic, prognostic and therapeutic applications. Semin Cancer Biol 2018;52:85-102. [Crossref] [PubMed]
- Hussein D, Dallol A, Quintas R, et al. Overlapping variants in the blood, tissues and cell lines for patients with intracranial meningiomas are predominant in stem cell-related genes. Heliyon 2020;6:e05632. [Crossref] [PubMed]
- Filbin MG, Tirosh I, Hovestadt V, et al. Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq. Science 2018;360:331-5. [Crossref] [PubMed]
- Magill ST, Vasudevan HN, Seo K, et al. Multiplatform genomic profiling and magnetic resonance imaging identify mechanisms underlying intratumor heterogeneity in meningioma. Nat Commun 2020;11:4803. [Crossref] [PubMed]
- Neftel C, Laffy J, Filbin MG, et al. An Integrative Model of Cellular States, Plasticity, and Genetics for Glioblastoma. Cell 2019;178:835-849.e21. [Crossref] [PubMed]
- Wang L, Babikir H, Müller S, et al. The Phenotypes of Proliferating Glioblastoma Cells Reside on a Single Axis of Variation. Cancer Discov 2019;9:1708-19. [Crossref] [PubMed]
- Khan I, Baeesa S, Bangash M, et al. Pleomorphism and drug resistant cancer stem cells are characteristic of aggressive primary meningioma cell lines. Cancer Cell Int 2017;17:72. [Crossref] [PubMed]
- Wang Z, Feng X, Li SC. SCDevDB: A Database for Insights Into Single-Cell Gene Expression Profiles During Human Developmental Processes. Front Genet 2019;10:903. [Crossref] [PubMed]
- Heiland DH, Staszewski O, Hirsch M, et al. Malignant Transformation of a Dysembryoplastic Neuroepithelial Tumor (DNET) Characterized by Genome-Wide Methylation Analysis. J Neuropathol Exp Neurol 2016;75:358-65. [Crossref] [PubMed]
- Matsumura N, Natsume A, Maeda S, et al. Malignant transformation of a dysembryoplastic neuroepithelial tumor verified by a shared copy number gain of the tyrosine kinase domain of FGFR1. Brain Tumor Pathol 2020;37:69-75. [Crossref] [PubMed]
- Kazemi-Sefat GE, Keramatipour M, Talebi S, et al. The importance of CDC27 in cancer: molecular pathology and clinical aspects. Cancer Cell Int 2021;21:160. [Crossref] [PubMed]
- Ben-David U, Amon A. Context is everything: aneuploidy in cancer. Nat Rev Genet 2020;21:44-62. [Crossref] [PubMed]
- Håvik AL, Bruland O, Myrseth E, et al. Genetic landscape of sporadic vestibular schwannoma. J Neurosurg 2018;128:911-22. [Crossref] [PubMed]
- Kohsaka S, Shukla N, Ameur N, et al. A recurrent neomorphic mutation in MYOD1 defines a clinically aggressive subset of embryonal rhabdomyosarcoma associated with PI3K-AKT pathway mutations. Nat Genet 2014;46:595-600. [Crossref] [PubMed]
- Wu R, Li Q, Wu F, et al. Comprehensive Analysis of CDC27 Related to Peritoneal Metastasis by Whole Exome Sequencing in Gastric Cancer. Onco Targets Ther 2020;13:3335-46. [Crossref] [PubMed]
- Wang DP, Gu LL, Xue Q, et al. CtBP2 promotes proliferation and reduces drug sensitivity in non-small cell lung cancer via the Wnt/β-catenin pathway. Neoplasma 2018;65:888-97. [Crossref] [PubMed]
- Kong S, Cao Y, Li X, et al. MiR-3116 sensitizes glioma cells to temozolomide by targeting FGFR1 and regulating the FGFR1/PI3K/AKT pathway. J Cell Mol Med 2020;24:4677-86. [Crossref] [PubMed]
- Mancarella C, Scotlandi K. IGF2BP3 From Physiology to Cancer: Novel Discoveries, Unsolved Issues, and Future Perspectives. Front Cell Dev Biol 2019;7:363. [Crossref] [PubMed]


