
Differences in gene expression between the primary and secondary inferior oblique overaction
Introduction
Inferior oblique overaction (IOOA) is very common in patients with strabismus. It is manifested by the overelevation of the eye in adduction and is frequently observed in superior oblique palsy (SOP), congenital esotropia, accommodative esotropia, and intermittent exotropia (1,2). In congenital esotropia, patients present clinically with a large angle esotropia which usually occurs within 6 months after birth and may be accompanied by inferior oblique overaction, dissociated vertical deviation, or nystagmus (3). The reasons for the IOOA in congenital esotropia remain unknown, and thus it is referred as primary IOOA. A study by Wilson et al reported that IOOA developed in 72% of congenital esotropia (1). In superior oblique palsy, the main clinical symptom is binocualr vertical misalignment. There is hypertropia of the affected eye in primary position, which increases when the eye is adducted or head tilted toward the affected eye (4). The annual incidence rate of superior oblique palsy in children was 3.4 cases per 100,000 people (5). The IOOA in SOP mainly results from hypofunction of the ipsilateral superior oblique muscle, and thus it is called secondary IOOA (6). Both congenital esotropia and SOP share the same symptom of IOOA; however, their visual perceptual states differ. In congenital esotropia, the fusion faculty is considered absent, and binocularity does not exist in these patients (3,6). However, in SOP, patients adopt a compensated head posture to help eliminate diplopia and obtain a normal retinal correspondence and good binocularity (4,7,8). The 4 cyclovertical muscles in our eyes (which comprise 2 superior and 2 inferior oblique muscles) are mainly involved in the vertical fusion of SOP, and patients with congenital SOP usually have a larger than normal vertical fusion amplitude. However, the cyclovertical muscles may not work in vertical fusion in congenital esotropia due to a loss of fusion.
Kono and Demer compared the cross-sectional size of inferior oblique (IO) muscles between SOP patients and normal subjects using a magnetic resonance imaging technique, but no significant differences were found (9). Additionally, in another study, the mean cross-sectional areas of the IO muscles from primary IOOA were also compared to those of normal control subjects, and similarly, no differences between the two groups were observed (10). This may suggest that the overacting IO muscles, whether in primary or secondary IOOA, do not present morphological hypertrophy.
In this study, we sought to identify the molecular differences between the primay and secondary overacting IO muscles. IO muscles were collected from patients who were diagnosed with congenital SOP or congenital esotropia and needed to undergo a weakening surgery of the inferior oblique muscle. Muscle samples from congenital esotropia patients were used for the control group. A transcriptome analysis of muscle tissues was conducted to help understand the different adaptive changes of the inferior oblique muscle at the subcellular and molecular levels.
Methods
Patient and tissue sample collection
The study was conducted between April 2021 to December 2021 at the Ophthalmology Department of Beijing Children’s Hospital, Capital Medical University. Inferior oblique muscles were obtained from patients who required a weakening procedure of the inferior oblique muscle. A total of 8 patients aged between 8 to 40 months were included in the study, 5 of whom had been diagnosed with congenital SOP and 3 of whom had been diagnosed with congenital esotropia. Patients with a history of other ocular diseases, previous eye surgery, and systemic diseases were excluded from the study. All the subjects received a complete ophthalmology examination, and all the surgeries were performed by the same pediatric ophthalmologist. The tissue samples harvested during the operation were snap-frozen in liquid nitrogen, and then stored at –80 ℃. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Ethics Committee of Beijing Children’s Hospital, Capital Medical University (No. 2016-42) and the parents or guardians of the children provided written informed consent.
RNA library preparation and transcriptome sequencing
The total ribonucleic acids (RNAs) were extracted from each extraocular muscle sample using TRIzol (Invitrogen) following standard procedures. The purity and integrity of the RNAs were assessed using a NanodropTM spectrophotometer and 1.5% agarose gel electrophoresis. The RNA quantification was carried out by Qubit 3.0 with a QubitTM RNA Broad Range Assay Kit (Life Technologies). Total RNA (2 µg) was used for library preparation with the KC-DigitalTM Stranded mRNA Library Prep Kit for Illumina® (catalog no. DR08502, Seqhealth Technology Co., Ltd. Wuhan, China). The enriched fragments (200 to 500 bp) were sequenced on Illumina Novaseq 6000.
Screening of DEGs
Raw-data processing was performed by removing the low-quality reads and reads contaminated with adaptor sequences through Trimoraic (version 0.36) (11). Next, the obtained clean reads were further analyzed with in-house scripts (Kangce Technology Co., Ltd., Wuhan, China) to eliminate amplification bias during the library preparation and sequencing (12,13). Subsequently, a standard RNA-sequencing procedure was performed. The sequencing reads were mapped to the human reference genome (hg38) using STAR software (version 2.5.3a) (14). The reads per kilobase million were calculated as a measure of expression value (15). The differentially expressed gene (DEG) analysis was performed in R language (version 3.6.3) using the DESeq2 R package (16).
Functional enrichment analysis
The Gene Ontology (GO) functional enrichment analyses, including the analyses of the biological process (BP), molecular function (MF), and cellular component (CC), were conducted by using R clusterProfiler package (version 3.14.3) (17). Then, the data were visualized by using ggplot 2 package (version 3.3.3) and both the upregulated and downregulated DEGs were entered separately.
GSEA
The gene set enrichment analysis (GSEA) of the whole transcriptome data was conducted via clusterProfiler (version 3.14.3) (17)to determine the Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathways. Annotated gene sets (c2.cp.v7.2.symbols.gmt) were selected for the GSEA analysis. The gene list input for the GSEA was ranked by values of log2 fold-change. The R package ggplot 2 (version 3.3.3) was used for data visualization.
PPI network analysis
To further analyze the protein-protein interactions (PPIs) between the corresponding genes, the DEGs were added to the Search Tool for the Retrieval of Interacting Genes (STRING; http://string-db.org) online database. The PPI network was then further analyzed and visualized using Cytoscape software (version 3.8.2). The Molecular Complex Detection (MCODE) plugin was used to identify significant subclusters. By default, we set the degree cutoff to 2, the K-score to 2, and the maximum depthto 100.
Statistical analysis
We screened the DEGs by R with DESeq2 R package and the cutoff parameters were P<0.05 and |log FC| >1. A t-test was used to compare the RNA-seq expression levels of interested genes between the two groups. GO and GSEA were performed for biological function analyses by using clusterProfiler package (version 3.14.3). An adjusted P value (P adj) <0.05 was considered statistically significant for GO analysis. The results of GSEA were considered significant when satisfying the criteria of P adj <0.05 and false discovery rate q<0.25.
Results
Identification of DEGs
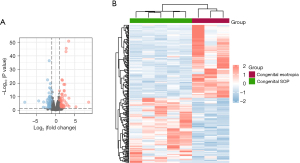
The inferior oblique muscles of both the congenital SOP and congenital esotropia patients were collected for RNA-sequencing. As a result, a total of 16,735 co-expressed genes were identified among the two groups. Using the screening criteria of P<0.05 and |log FC| >1, we identified 221 DEGs, including 104 upregulated genes and 117 downregulated genes. The volcano plots and heatmap show the different gene expression patterns (see Figure 1A,1B). The expression levels of the 4 DEGs related to muscle contraction force were analyzed using the RNA-sequencing data (see Figure 2).
Analysis of GO enrichment
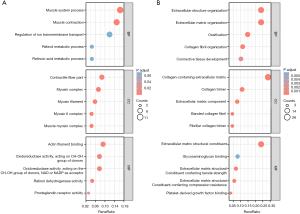
To further examine the function of the DEGs, GO enrichment analyses of the upregulated and downregulated genes were conducted, and a total of 105 GO terms were identified. For the BP analysis, the upregulated DEGs were significantly enriched in the muscle system process and muscle contraction, while the downregulated DEGs were significantly enriched in extracellular structure organization, extracellular matrix (ECM) organization, ossification, collagen fibril organization, and connective tissue development. For the CC analysis, the upregulated genes were primarily related to the contractile fiber part, myosin complex, myosin filament, myosin II complex, and muscle myosin complex, while the downregulated genes were primarily related to collagen containing the ECM, the collagen trimer, the ECM component, the banded collagen fibril, and the fibrillar collagen trimer. For the MF analysis, the upregulated genes were mostly associated with actin filament binding, oxidoreductase activity, retinol dehydrogenase activity, and prostaglandin receptor activity, while the downregulated genes were mostly associated with the ECM structural constituent, glycosaminoglycan binding, and platelet-derived growth factor binding (see Figure 3). Overall, the DEGs mainly related to muscle contraction and muscle process were significantly upregulated in the inferior oblique muscle of SOP, while the DEGs related to the ECM components and metabolism were downregulated.
Pathways identified by GSEA
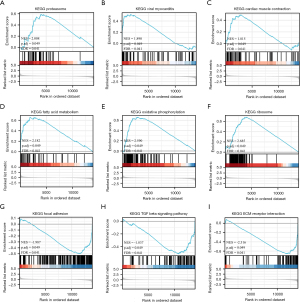
To identify the related pathways, a GSEA was conducted. In total, 18 differently regulated KEGG pathways were identified (see Table 1), among which, 14 were upregulated, and 4 were downregulated. Some of the most interesting pathways included proteasome (NES =2.008, P adj =0.049), viral myocarditis (NES =1.890, P adj =0.049), cardiac muscle contraction (NES =1.815, P adj =0.049), fatty acid metabolism (NES =2.182, P adj =0.049), oxidative phosphorylation (NES =2.590, P adj =0.049), ribosome (NES =2.685, P adj =0.049), focal adhesion (NES =–1.987, P adj =0.049), the transforming growth factor beta signaling pathway (NES =–1.837, P adj =0.049), and ECM receptor interaction (NES =–2.316, P adj =0.049) (see Figure 4). Significance was defined as P adj <0.05 and q<0.25. NES refers to the normalized enrichment score, and the q value is equivalent to the false discovery rate.
Table 1
| KEGG ID | setSize | NES | P value | P adjusted value | Q value |
|---|---|---|---|---|---|
| Ribosome | 86 | 2.685401597 | 0.002739726 | 0.048600106 | 0.04085871 |
| Oxidative phosphorylation | 91 | 2.589709656 | 0.00280112 | 0.048600106 | 0.04085871 |
| Parkinson’s disease | 92 | 2.312832855 | 0.002762431 | 0.048600106 | 0.04085871 |
| Antigen processing and presentation | 52 | 2.230255219 | 0.002610966 | 0.048600106 | 0.04085871 |
| Drug metabolism cytochrome p450 | 31 | 2.207146113 | 0.002439024 | 0.048600106 | 0.04085871 |
| Fatty acid metabolism | 36 | 2.182363948 | 0.002493766 | 0.048600106 | 0.04085871 |
| Allograft rejection | 21 | 2.163845304 | 0.002331002 | 0.048600106 | 0.04085871 |
| Graft versus host disease | 20 | 2.120080024 | 0.002369668 | 0.048600106 | 0.04085871 |
| Type I diabetes mellitus | 24 | 2.034925493 | 0.002457002 | 0.048600106 | 0.04085871 |
| Alzheimer’s disease | 131 | 2.023485206 | 0.003134796 | 0.048600106 | 0.04085871 |
| Proteasome | 40 | 2.008203865 | 0.002493766 | 0.048600106 | 0.04085871 |
| Viral myocarditis | 61 | 1.890222478 | 0.002695418 | 0.048600106 | 0.04085871 |
| Huntington’s disease | 144 | 1.843588014 | 0.003067485 | 0.048600106 | 0.04085871 |
| Cardiac muscle contraction | 62 | 1.814980004 | 0.002673797 | 0.048600106 | 0.04085871 |
| Axon guidance | 104 | –1.691815181 | 0.002989537 | 0.048600106 | 0.04085871 |
| Transforming growth factor beta signaling pathway | 70 | –1.836746472 | 0.003169572 | 0.048600106 | 0.04085871 |
| Focal adhesion | 182 | –1.986682979 | 0.001436782 | 0.048600106 | 0.04085871 |
| ECM receptor interaction | 69 | –2.315790338 | 0.0016 | 0.048600106 | 0.04085871 |
KEGG, Kyoto Encyclopedia of Genes and Genomes; NES, normalized enrichment score; GSEA, gene set enrichment analysis; ECM, extracellular matrix.
PPI network construction and module identification
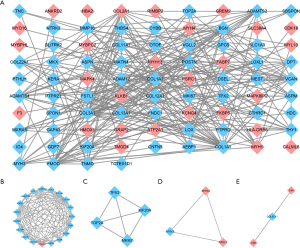
The DEGs were uploaded to the STRING online database to identify known and predicted PPIs. Next, we constructed the PPI network using Cytoscape software. As a result, 102 nodes and 295 edges were obtained with a minimum interaction score of 0.4 (see Figure 5A). A total of 4 cluster modules were identified by MCODE (see Figure 5B-5E).
Discussion
In a normal situation, there are two mechanisms for the maintenance of binocular alignment, including vergence adaption and muscle length adaption. Vergence adaption comprises “fast” fusional vergence, which is invoked by retinal image disparity, and “slow” tonic vergence, which can be sustained for minutes or hours (18). Both fusional vergence and tonic vergence are present in congenital SOP for vertical fusion under compensatory head tilting; however, these may be absent in congenital esotropia due to an absence of fusion.
Studies have reported that fundus extortion in congenital esotropia occurs before the development of IOOA (overelevation in adduction) (19,20). Thus, it has been speculated that fundus extortion may result from loss of fusion other than IOOA in congenital esotropia (20,21). In patients with congenital SOP, there is also extortion of the eye which is secondary to a decreased stimulation to the superior oblique muscle. However, SOP patients always engage in compensatory head tilting to keep the eyes aligned in vertical direction (22). Under this vertical fusion, a chronic and increased innervation to the ipsilateral inferior oblique muscle occurs, which results in the strengthening of the inferior oblique muscle in SOP.
In this study, we compared the gene expression profiles of the 2 diseases and identified 221 DEGs (|log FC| >1, P<0.05), among which 104 were upregulated, and 117 were downregulated. Several bioinformatics methods were then used to conduct further analysis. To find the different adaptive changes in gene expression resulting from the vertical fusion in SOP, we mainly focused on the upregulated genes and related pathways in the following analyses.
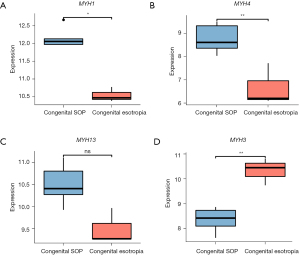
The GO enrichment analysis showed that the upregulated genes were mainly related to muscle contraction (ATP2A1, KCNJ3, MYBPC2, MYH1, MYH4, MYH13, and ANKRD2) and muscle system processes (ATP2A1, HMOX1, KCNJ3, MYBPC2, MYH1, MYH4, and MYH13). As is well known, the myosin heavy chain (MyHC) is very important for contraction force generation. To date, a total of 11 MyHC isoforms have been identified in mammalian skeletal muscles, all of which have different kinetic profiles (23,24). Myofibers containing different MyHC isoforms are divided into the following two main types: (I) the fast-twitch type; and (II) the slow-twitch type. It is well known that fast-twitch fibers produce higher contraction force and speed than slow-twitch fibers. In our study, 4 different MyHC (MYH) isoforms were found to be significantly differentially expressed, including 3 fast-twitch types (i.e., MyHC1 (MYH1, MyHC-2X) (25,26), MyHC4 (MYH4, MyHC-2B) (27), and MyHC113 (MYH13, MyHC-EO) (28), and 1 slow-twitch type (i.e., MyHC3 (MYH3, MyHC-emb) (29,30). All 3 fast-twitch MyHC isoforms were upregulated, and the only 1 slow-twitch isoform was downregulated. It is known that an increase in muscle’s cross-sectional area, increase in innervation, and changes in muscle fiber types can cause an increase in a muscle’s contractile force. The fast isoforms of MyHC produce stronger and faster muscle contractions than slow isoforms of MyHC. Thus, it was inferred that the changes in expression levels of these MyHC isoforms may play a key role in the strengthening of inferior oblique muscle in SOP.
The enriched KEGG pathways from the GSEA revealed that viral myocarditis, cardiac muscle contraction, ribosome, oxidative phosphorylation, fatty acid metabolism, and proteasome may play significant roles in muscle adaption. As is well known, ribosomes are essential for protein synthesis in cells. Generally, the number of ribosomes determines the rate of protein synthesis per cell (31). Studies have shown that ribosome biogenesis is associated with increased myofibrillar muscle protein synthesis and muscle hypertrophy (32-34). The metabolism pathways of oxidative phosphorylation and fatty acid metabolism were upregulated, they are important biochemical processes for energy producing. Proteasomes are responsible for the maintenance of protein homeostasis in cells, and degrade both normal and damaged proteins (35).
A previous study reported that, the myogenesis-related genes MYOD1 and myogenin were higher in the overacting inferior oblique muscle than the normal controls. Therefore, the author inferred that there were more activated satellite cells in the overacting IO muscles (10). In our study, we did not find any difference in the expression of specific markers for activated satellite cells, as the control samples in our study were not normal controls. However, the results may indicate that there is no difference in myogenesis-related genes between the secondary and primary overacting inferior oblique muscles.
Usually, MyHC isoforms are regarded as molecular markers of different muscle fiber types. Studies on MyHC isoforms have focused primarily on the transition of different MyHC isoforms (slow-fast transitions). The main factors that can regulate the muscle phenotype include pattern of neural stimulation, loading and unloading, oxygen and substrate availability and hormonal signals (36). However, the vast majority of studies were based on mammalian limb and truck muscles other than extraocular muscles. This is because that the extraocular muscles are more complex in MyHC composition, even singer extraocular muscle fiber may co-express different MyHC isoforms which vary along the extent of the muscle fiber (23). In spite of the findings in this study, more research is still needed for the underlying pathogenic mechanism and to even discover a potential direction for the treatment.
This study had a number of limitations. First, in this study, only the distal portion of the inferior oblique muscles were available during surgery; thus, the results do not represent the complex changes of the whole inferior oblique muscle. Second, it is very difficult to obtain muscle samples from healthy children, as normal muscles are almost never removed. This limits our ability to further interpret the downregulated DEGs in SOP compared to congenital esotropia. Finally, the actual molecular mechanisms that underlie this process remain unclear; thus, the further experimental validation of our results is required.
Conclusions
To date, no previous studies appear to have examined the differences of transcriptional profiling in primary and secondary overacting inferior oblique muscles. In our study, we found that several genes related to fast isoforms of MyHC in the inferior oblique muscle of SOP were upregulated and this may contribute to its strengthening. Pathways, such as viral myocarditis, cardiac muscle contraction, ribosome, oxidative phosphorylation, fatty acid metabolism, and proteasome, might be involved in the pathological changes of inferior oblique muscle in SOP. Further research needs to be conducted to elucidate the precise molecular mechanism.
Acknowledgments
Funding: The work was supported by funding from the National Natural Science Foundation of China (No. 81670883).
Footnote
Data Sharing Statement: Available at https://tp.amegroups.com/article/view/10.21037/tp-22-98/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-22-98/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work, including ensuring that any questions related to the accuracy or integrity of any part of the work have been appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of the Beijing Children’s Hospital, Capital Medical University (No. 2016-42) and the parents or guardians of the children provided written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wilson ME, Parks MM. Primary inferior oblique overaction in congenital esotropia, accommodative esotropia, and intermittent exotropia. Ophthalmology 1989;96:950-5; discussion 956-7. [Crossref] [PubMed]
- Meyer E, Ludatscher RM, Zonis S. Primary and secondary overacting inferior oblique muscles: an ultrastructural study. Br J Ophthalmol 1984;68:416-20. [Crossref] [PubMed]
- Nelson LB, Wagner RS, Simon JW, et al. Congenital esotropia. Surv Ophthalmol 1987;31:363-83. [Crossref] [PubMed]
- Chang MY, Coleman AL, Tseng VL, et al. Surgical interventions for vertical strabismus in superior oblique palsy. Cochrane Database Syst Rev 2017;11:CD012447. [Crossref] [PubMed]
- Tollefson MM, Mohney BG, Diehl NN, et al. Incidence and types of childhood hypertropia: a population-based study. Ophthalmology 2006;113:1142-5. [Crossref] [PubMed]
- Ozsoy E, Gunduz A, Ozturk E. Inferior Oblique Muscle Overaction: Clinical Features and Surgical Management. J Ophthalmol 2019;2019:9713189. [Crossref] [PubMed]
- Jiang L, Demer JL. Magnetic resonance imaging of the functional anatomy of the inferior rectus muscle in superior oblique muscle palsy. Ophthalmology 2008;115:2079-86. [Crossref] [PubMed]
- Farid MF, Anany M, Abdelshafy M. Surgical outcomes of three different weakening procedures of inferior oblique muscle in the treatment of unilateral superior oblique palsy. BMC Ophthalmol 2020;20:298. [Crossref] [PubMed]
- Kono R, Demer JL. Magnetic resonance imaging of the functional anatomy of the inferior oblique muscle in superior oblique palsy. Ophthalmology 2003;110:1219-29. [Crossref] [PubMed]
- Antunes-Foschini RM, Ramalho FS, Ramalho LN, et al. Increased frequency of activated satellite cells in overacting inferior oblique muscles from humans. Invest Ophthalmol Vis Sci 2006;47:3360-5. [Crossref] [PubMed]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 2014;30:2114-20. [Crossref] [PubMed]
- Shugay M, Britanova OV, Merzlyak EM, et al. Towards error-free profiling of immune repertoires. Nat Methods 2014;11:653-5. [Crossref] [PubMed]
- Ma KY, He C, Wendel BS, et al. Immune Repertoire Sequencing Using Molecular Identifiers Enables Accurate Clonality Discovery and Clone Size Quantification. Front Immunol 2018;9:33. [Crossref] [PubMed]
- Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15-21. [Crossref] [PubMed]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923-30. [Crossref] [PubMed]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [Crossref] [PubMed]
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. [Crossref] [PubMed]
- Guyton DL. The 10th Bielschowsky Lecture. Changes in strabismus over time: the roles of vergence tonus and muscle length adaptation. Binocul Vis Strabismus Q 2006;21:81-92. [PubMed]
- Eustis HS, Nussdorf JD. Inferior oblique overaction in infantile esotropia: fundus extorsion as a predictive sign. J Pediatr Ophthalmol Strabismus 1996;33:85-8. [Crossref] [PubMed]
- Kushner BJ. Multiple mechanisms of extraocular muscle "overaction". Arch Ophthalmol 2006;124:680-8. [Crossref] [PubMed]
- Lagstein O, Guyton DL. Inferior Oblique Muscle "Overaction" Caused by Inferior Oblique Muscle Shortening, Not by Hypertonicity. J Pediatr Ophthalmol Strabismus 2022;59:28-34. [Crossref] [PubMed]
- Kline LB, Demer JL, Vaphiades MS, et al. Disorders of the Fourth Cranial Nerve. J Neuroophthalmol 2021;41:176-93. [Crossref] [PubMed]
- Hoh JFY. Myosin heavy chains in extraocular muscle fibres: Distribution, regulation and function. Acta Physiol (Oxf) 2021;231:e13535. [Crossref] [PubMed]
- Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev 2011;91:1447-531. [Crossref] [PubMed]
- LaFramboise WA, Daood MJ, Guthrie RD, et al. Electrophoretic separation and immunological identification of type 2X myosin heavy chain in rat skeletal muscle. Biochim Biophys Acta 1990;1035:109-12. [Crossref] [PubMed]
- DeNardi C, Ausoni S, Moretti P, et al. Type 2X-myosin heavy chain is coded by a muscle fiber type-specific and developmentally regulated gene. J Cell Biol 1993;123:823-35. [Crossref] [PubMed]
- Rinaldi C, Haddad F, Bodell PW, et al. Intergenic bidirectional promoter and cooperative regulation of the IIx and IIb MHC genes in fast skeletal muscle. Am J Physiol Regul Integr Comp Physiol 2008;295:R208-18. [Crossref] [PubMed]
- Briggs MM, Schachat F. Early specialization of the superfast myosin in extraocular and laryngeal muscles. J Exp Biol 2000;203:2485-94. [Crossref] [PubMed]
- Lyons GE, Ontell M, Cox R, et al. The expression of myosin genes in developing skeletal muscle in the mouse embryo. J Cell Biol 1990;111:1465-76. [Crossref] [PubMed]
- Condon K, Silberstein L, Blau HM, et al. Development of muscle fiber types in the prenatal rat hindlimb. Dev Biol 1990;138:256-74. [Crossref] [PubMed]
- Paul BJ, Ross W, Gaal T, et al. rRNA transcription in Escherichia coli. Annu Rev Genet 2004;38:749-70. [Crossref] [PubMed]
- Brook MS, Wilkinson DJ, Mitchell WK, et al. A novel D2O tracer method to quantify RNA turnover as a biomarker of de novo ribosomal biogenesis, in vitro, in animal models, and in human skeletal muscle. Am J Physiol Endocrinol Metab 2017;313:E681-9. [Crossref] [PubMed]
- Kim HG, Guo B, Nader GA. Regulation of Ribosome Biogenesis During Skeletal Muscle Hypertrophy. Exerc Sport Sci Rev 2019;47:91-7. [Crossref] [PubMed]
- Wen Y, Alimov AP, McCarthy JJ. Ribosome Biogenesis is Necessary for Skeletal Muscle Hypertrophy. Exerc Sport Sci Rev 2016;44:110-5. [Crossref] [PubMed]
- Thibaudeau TA, Smith DM. A Practical Review of Proteasome Pharmacology. Pharmacol Rev 2019;71:170-97. [Crossref] [PubMed]
- Blaauw B, Schiaffino S, Reggiani C. Mechanisms modulating skeletal muscle phenotype. Compr Physiol 2013;3:1645-87. [Crossref] [PubMed]


