
A de novo pure 21q22.3 deletion in a 9-year-old boy with buried penis: a case report and literature review
Introduction
Trisomy is the most common abnormality of chromosome 21. Whereas fetuses with complete 21q deletion die before or shortly after birth, cases with partial deletion of chromosome 21 have a better survival expectancy. While a very rare condition (<1/1,000,000), partial deletion of chromosome 21q can affect a variety of human systems due to the gene dosage deficiency (1). The diagnosis and treatment of 21q22.3 deletion is difficult and easy to misdiagnose, which is worthy of clinicians’ attention. Typical clinical symptoms include intrauterine growth retardation, microcephaly, a low auricle, heart defects, seizures, intellectual disability, mental and language disorders, and dysplasia of the corpus callosum (2). While most 21q deletions are associated with other chromosomal abnormalities, the patient in the present study has a rare pure 21q22.3 deletion. At present, the most comprehensive studies about 21q deletion are from Roberson et al. (3) and Lyle et al. (4), who discussed 11 and 10 cases of partial 21q deletion respectively. The size of the deletions ranged from 1.48 to 21.06 Mb, and all cases were unique, without an identical breakpoint. Roberson et al. (3) summarized the chromosome 21q deletion breakpoints for 36 cases and found that 20 involved aneuploidy and/or a translocation on a chromosome other than 21. Lyle et al. divided 21q into three regions according to the severity of phenotypes and found region 1 (centromere to 31.2 Mb), region 2 (31.2 Mb to 36 Mb), and region 3 (36 Mb to telomere) were respectively associated with mild, moderate, and severe phenotypes. Deletions in region 1 tended to be large. Errichiello et al. (1) later proposed to delimit region 1 to two subregions. The deleted fragment described in this article was in region 3, which is reported as the most frequently altered area.
Partial deletion of chromosome 21 is heterogeneous in terms of phenotypic severity. We report a rare case of a boy aged 9 with concurrent intellectual disability, brain anomalies, and gonadal dysgenesis resulting from 21q22.3 partial deletion. We compared the deletion regions in cases listing brain anomalies and gonadal dysgenesis from the literature and DECIPHER, and summarized two microdeletion regions that may be related to gonadal and brain development.
We present the following case in accordance with the CARE reporting checklist (available at https://dx.doi.org/10.21037/tp-21-377).
Case presentation
The case concerns a 9-year-old Chinese boy of Han ethnicity who was the product of the first pregnancy of unrelated healthy parents and was delivered by caesarean section at full term with a birth weight of 3.1 kg. His mother was 26 years old, 156 cm in height and weighed 57 kg, and her prenatal history revealed no evidence of teratogen or drug exposure. His father was 33 years old at the time of his birth and was 166 cm tall and weighed 51 kg. The developmental milestones of raising the head, turning over, and sitting up were achieved at normal times by the child. He walked at the age of two and started speaking at three and a half years of age. While there was no history of epilepsy, he occasionally fell down and was unable to run due to the instability and poor coordination of his limbs during exercise. His intelligence lagged behind peers, and he was poor at language expression, interpersonal communication, learning, calculating, and writing. He had a low attention span and often laughed involuntarily. His eyes had astigmatism and myopia and his penis was short and could not be exposed (Figure 1).
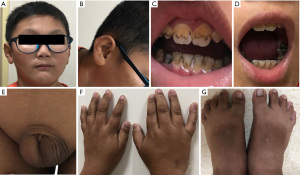
On physical examination, the boy had a regular pulse (86 beats per min), blood pressure (100/78 mmHg), height of 129 cm (which was less than others of the same age and sex by 1 SD), weight of 32 kg, BMI of 19.22 kg/m2, head circumference of 51 cm, fingertip distance of 130 cm, upper body height of 65 cm, and lower body height of 64 cm.
He had multiple dental caries, a low hairline, a low auricle, dark skin (especially at the elbow and interphalangeal joints) and his hands and toes showed oblique deformity. Cardiopulmonary and abdominal examination showed no abnormality. His pubic hair and genitalia were at Tanner stage I, the penis was not exposed, and the testes could not be touched. The penile length from the tip of the glans penis to the suprapubic bone was 1.5 cm. The patient had a poor sense of balance and could not complete a both-hands alternating movement test, heel-knee-tibia test, or finger-nose test. The muscle strength and tension were normal, and the pathological sign was negative.
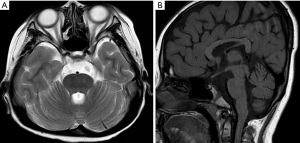
An auxiliary examination (Infant-Junior Middle School Student’s Ability of Social Life Scale) (5) showed moderate mental retardation. The following tests were all normal: liver and kidney function, blood lipid, myocardial enzyme, ACTH and cortisol levels and rhythm, thyroid function, ECG, chest X-ray, adrenal CT, color Doppler ultrasound of heart, abdomen, and urine. IGF-1 was 68.2 ng/mL (74–388 ng/mL). Growth hormone could not be stimulated by insulin hypoglycemia test or levodopa stimulation test (Table 1), indicating a deficiency. Gonadal hormones analysis showed LH 0.05 IU/L (0.57–12.07 IU/L), FSH 0.32 IU/L (0.95–11.95 IU/L), estradiol 37.0 pmol/L (40–161 pmol /L), prolactin 13.9 ng/mL (3.4–19.2 ng/mL), and testosterone 0.44 nmol/L (6.5–33 nmol/L). These levels suggested a deficiency of LH and FSH secreted by the pituitary gland. EEG showed mild abnormality, and testicular color Doppler ultrasound showed that the left testis was approximately 13 mm × 7 mm × 11 mm, the right testis was 15 mm × 8 mm × 12 mm, and the volume of the bilateral testis was small. The bone age was about 8 years old, and pituitary MRI showed the prepontine cistern and annular cistern to be widened and the pituitary gland to be thinned and hypoplastic (Figure 2).
Table 1
| Test | Factor | 0 min | 15 min | 30 min | 60 min | 90 min | 120 min |
|---|---|---|---|---|---|---|---|
| Insulin hypoglycemia test | GH (ng/mL) | 0.03 | 0.03 | 0.355 | 1.08 | 0.25 | 0.103 |
| BG (mmol/L) | 4.5 | 1.7 | 2.1 | 3.9 | 4.6 | 4.6 | |
| Levodopa stimulation test | GH (ng/mL) | 0.03 | 0.03 | 0.03 | 0.03 | 0.45 |
GH, growth hormone; BG, blood glucose.
After informed consent was obtained, 5ml of peripheral blood was collected from the patient and his parents. The process of whole exome sequencing was performed by WeHealth Biomedical Technology Co., Ltd. (Shanghai, China), and all genomic DNA was extracted using a commercial kit (TIANGEN, China). The quantity/quality of DNA was assessed using a Onedrop OD1000 spectrophotometer and by agarose gel electrophoresis. Exome capture was performed with xGen Exome Research Panel v1.0 (Integrated DNA Technologies, Inc., USA) and 150 bp paired-end sequencing was executed using the Illumina HiSeq4000 platform (San Diego, CA, USA).
For whole-exome sequencing bioinformatic analysis, the raw reads were aligned by the sequencing company using the Burrows-Wheeler Aligner and SAMtools. After removing duplicates from the sorted alignment using Picard, variants were called using the Genome Analysis Toolkit (GATK v3.70) pipeline.
For low depth whole genome sequencing, low-quality reads were removed by Trimmomatic v0.32. Sequences were aligned to the hg19 reference genome by Bowtie 2 v2.3.4.3 and duplicates reads were removed by Picard. A CNVkit v0.9.6 software toolkit was used to infer and visualize the copy number.
Based on the analysis of genome copy number variation by low depth whole genome sequencing technology, the patient was found to have a deletion of 4.5Mb on chromosome 21 (Figure 3) in the location of chr21: 43531239-48119895 (GRCh37/hg19). The karyotype was 46, XY, del (6) (q22.3), which involves 124 genes, including 18 known pathogenic genes in OMIM (including WDR4, SIK1, and TSPEAR) and 106 non-pathogenic genes (including DIP2A, S100B, and PRMT2). By low depth whole genome sequencing detection, there was no abnormality in his parents’ 21q22.3 region, confirming a de novo alteration.
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the patient’ parents for publication of this case report and accompanying images. A copy of the written consent is available for review by the editorial office of this journal.
Discussion
Comparing phenotypes between patients with 21q deletion is difficult. Most locations of the missing fragments are different, and 21q partial deletion is always combined with other abnormalities, such as ring chromosome, gene duplication and dystopy, and deletion of other chromosomes. Sgardioli et al. (7) reported a female characterized by mild deformities of the face and limbs, mental retardation, seizures, and mitral valve prolapse. Genetic testing found a terminal 4.7Mb deletion at the 21q22.3 region (43310796-48097372 GRCh37/hg19) and duplication at the 20q region. While this deletion region overlaps with our patient in most areas, the brain morphology and reproductive system of the Sgardioli et al. patient were normal, suggesting chromosome 20q duplication may interfere with the phenotype.
We searched for the genome location 21q22.3 in the DECIPHER database (/www.deciphergenomics.org) and found 142 cases with deletion in 21q22.3 region, and the phenotype was described in 103 cases. After filtering out 42 cases (40.8%) which carried concurrent mutations on some other chromosome regions, among the remaining 61 cases, 60 (detailed information in Table S1) showed an overlapped deletion region with the present patient. Notably, among them, 10 cases showed the 16.49 Mb deletion of chr21:31578129-48119895 (GRCh37/hg19), and 25 cases showed the 5.59 Mb deletion of chr21:42478130-48119895 (GRCh37/hg19), suggesting that these were two commonly deleted regions of pure partial 21q. The phenotypes of these patient’s varied greatly, ranging from mild to severe, even among patients carrying the same deletion region. So it is difficult to identify the 21q22.3 deletion in the case. However, patients with neurological and gonadal dysplasia are at increased risk of chromosome 21 abnormalities, attention should be paid to screening genes and chromosomes.
We focused on the 21q22.3 deletion-related abnormality in the genital and central nervous systems, as our patient had a buried penis, small testes and brain anomalies. We cannot explain this phenotype by a specific gene since the relationship between genotype and genital abnormality is still not well defined. Chen et al. (8) reported a male with 21q deletion (42543932-48119895) who showed a concealed penis and corpus callosum dysgenesis, while Oegema et al. (9) reported a case of cryptorchidism whose 21q deleted in chr38131848-42180291. The DECIPHER database contains only a few 21q deletion patients with a genital abnormity of decreased testicular size, oligospermia, delayed publication and/or cryptorchidism. Zeng et al. (10) reported a female with premature ovarian insufficiency with 21q deletion (14539866-28673235) and speculated it could result from the haploinsufficiency of some dosage-sensitive genes or genes with unclear function, or from environmental factors. We hypothesize that a combination of genetic and environmental factors resulted in the buried penis and small testes phenotypes of the patient in the present study.
Deletion of 21q is related to changes in human brain morphogenesis. Dose-sensitive genes in this region contribute to cortical development, and deletion can result in cortical dysplasia (11,12). Our patient had a widened prepontine cistern and annular cistern, hypoplasia of the pituitary gland, and deficiency of growth hormone and gonadal hormone. Ruiz-Botero and Pachajoa (13) reported on a Colombian girl of mixed race with corpus callosum hypoplasia and intellectual and mental disorders, and genetic testing found the deletion of 3.608 Mb (44482408-48090317) on chromosome 21q22.3 and a duplication on chromosome 7q. Valetto et al. (14) reported a boy with 21q deletion located at 38791571-43792093(GRCh37/hg19), who also suffered from hypoplasia of pituitary and corpus callosum. The overlapped region between this boy and our patient was very small (43531239-43792093), and no gene concerned with brain morphological changes was found. Oegema et al. (9) reported two patients with deletion of chr21:38131848-42180291 and chr21:36424426-40654602, respectively, and although their deleted regions did not overlap with our patient (Table 2) (2,3,8,9,14,15), they were also found to have hypoplasia of the corpus callosum. Therefore, we speculate that there are multiple gene loci regulating brain development on the long arm of chromosome 21.
Table 2
| Variable | The present patient | Chen et al. patient | Guion-Almeida et al. patient | Roberson et al. GM09868 | Briegel et al. patient | Valetto et al. patient | Oegema et al. patient 1 | Oegema et al. patient 2 |
|---|---|---|---|---|---|---|---|---|
| Breakpoint | 43531239-48119897 | 42543932-48119895 | 46625055-46884297 | 35677518-46921373 | 43945335-48097372 | 38791571-43792093 | 38131848-42180291 | 36424426-40654602 |
| Deletion size (Mb) | 4.5 | 5.7 | 219 | 11.2 | 4.5 | 4.9 | 4.1 | 4.2 |
| Sex | M | M | F | F | F | M | M | F |
| Height | −1 SD | −1 SD | −2 SD | −3.6 SD | −3 SD | −3 SD | ||
| Microcephaly | N | Y | Y | Y | Y | Y | ||
| Hypertelorism | N | Y | Y | N | ||||
| Large ears | N | Y | Y | Y | Y | Y | ||
| Broad nasal bridge | N | Y | Y | Y | Y | Y | ||
| Abnormal vision | Y | N | ||||||
| Short stature | Y | Y | Y | Y | Y | Y | Y | |
| Distal limbs abnormalities | Y | N | Y | Y | Y | Y | ||
| Carious teeth | Y | N | Y | Y | ||||
| Epilepsy/seizures | N | Y | Y | Y | Y | |||
| Speech delay | Y | Y | Y | Y | Y | |||
| Cardiac anomaly | N | Y | Y | N | Y | Y | ||
| Mental retardation | Y | N | Y | Y | Y | |||
| Intellectual retardation | Y | Y | Y | Y | Y | Y | ||
| Balance disorder | Y | N | Y | |||||
| Brain imaging | Widened prepontine cistern and annular cistern, thinned and hypoplastic pituitary gland | Corpus callosum dysgenesis, colpocephaly, ventriculomegaly, microcephaly | Sphenoethmoidal encephalocele, callosal agenesis; anterior pituitary was not visualized. | Hypoplastic of corpus callosum and pituitary stalk | Small frontal lobes, a thin corpus callosum and brain stem | Underdeveloped frontal gyri, thin corpus callosum and brain stem, loss of periventricular white matter ventricles | ||
| Genital abnormality | Concealed penis | Concealed penis | Cryptorchidism |
We also compared the deletion region in cases with brain anomalies and/or gonadal dysgenesis reported in the literature and DECIPHER (Figure 4 and Table 2) (2,3,8,9,14,15). As seen in Table 2, the most frequent phenotypes are of short stature, and have intellectual retardation, speech delay, epilepsy/seizures, distal limbs abnormalities, and a broad nasal bridge. As shown in Figure 4, most of these deletion regions were overlapped, and two recurrent microdeletion regions were identified, which may be related to gonadal and brain development. We summarized two overlapped microdeletion regions from these cases that may be related to gonadal and brain development and found 21q22.3 microdeletion 1 (chr21:43531239-43792093) and 21q22.3 microdeletion 2 (chr21:46625055-46884297) harbor seven genes and 10 genes respectively (Table 3). In 21q22.3 microdeletion region 1, gene ABCG1 is highly expressed in the brain and plays a critical role in cerebrovascular function (16-18). Studies have shown that loss of ABCG1 results in a chronic inflammatory response and endothelial cell injury and dysfunction. ADARB1 is a gene related to brain development in 21q22.3 microdeletion 2, and its mutation can lead to autosomal recessive neurodevelopmental disorders with hypotonia, microcephaly, and epilepsy (19). We did not find any genes associated with gonadal dysgenesis in these two microdeletion regions.
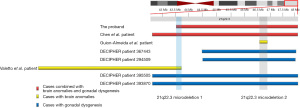
Table 3
| Variable | 21q22.3 microdeletion 1 | 21q22.3 microdeletion 2 |
|---|---|---|
| Location | chr21:43531239-43792093 (GRCh37) | chr21:46625055-46884297 (GRCh37) |
| Genes | ABCG1, RNA5SP492, TFF1, TFF2, TFF3, TMPRSS3, UMODL1 | ADARB1, COL18A1, COL18A1-AS1, COL18A1-AS2, LINC00205, LINC00315, LINC00316, LINC00334, MTCO1P3, POFUT2 |
Of the other genes in our patient’s deleted region, PRMT2 inhibits NF-kappaB-dependent transcription and promotes apoptosis (20). Abnormal regulation of PRMT2 protein, part of the arginine methyltransferase family, produced transcripts for chromatin-remodeling enzymes associated with reproductive system disease and cancer in a baboon endometriosis model (21). Abnormalities of the WDR4 gene cause microcephaly, growth deficiency, seizures, and brain malformations, and mutation of the TSPEAR gene may result in ectodermal dysplasia. DIP2A and S100B in the 21q22.3 region were reported to be associated with autism spectrum disorder (6), and DIP2A was also shown to be related to dyslexia (22). SIK1 mutations generate variant and truncated SIK1 proteins that are associated with severe developmental epilepsy (23), and compared with the wild type, average neurite length and number were significantly reduced in human mutant SIK1 neurons, and the expression of synaptic activity response element genes decreased, causing epilepsy in some cases (24). However, the present case showed no signs of epilepsy. The dosage sensitivity of the deleted genes is not clear, and many of the deleted genes do not have a known function.
Partial 21q deletion is often accompanied by mental and psychological abnormalities. Our patient had difficulty in communication, and often laughed involuntarily, suggesting a mental disorder. However, mental disorders are difficult to diagnose in childhood because they may be masked by intellectual retardation and other phenotypes, so the actual prevalence may be underestimated. Therefore, we suggest regular screening for psychiatric disorders in affected children (15).
In conclusion, the similar abnormalities (hypoplasia of the corpus callosum) in different patients with non-overlapping deletion regions suggests there are multiple gene loci regulating brain development on 21q. Microdeletion 1 (chr21:43531239-43792093) and microdeletion 2 (chr21:46625055-46884297) may be related to gonadal and brain development in 21q22.3. Chr21:31578129-48119895 and chr21:42478130-48119895 are two commonly deleted regions of pure partial 21q, and the same deletion has different phenotypes in DECIPHER cases suggesting the heterogeneity of 21q deletion. The relationship between phenotype and genotype requires further study, and more cases and molecular mechanism research are needed to clarify the effect of 21q deletion.
Acknowledgments
The authors are deeply grateful to the patient and his parents for their support. This study made use of data generated by the DECIPHER community, and a full list of centres who contributed to the generation of this data is available from
Funding: This study received the support of the major research and development program of Hainan Province (nos. ZDYF2019156).
Footnote
Reporting Checklist: The authors have completed the CARE reporting checklist. Available at https://dx.doi.org/10.21037/tp-21-377
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tp-21-377). All authors reported that this study received the support of the major research and development program of Hainan Province (nos. ZDYF2019156), and received the support of Hainan Province Clinical Medical Center, and this study made use of data generated by the DECIPHER community. The authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee(s) and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the patient’ parents for publication of this case report and accompanying images. A copy of the written consent is available for review by the editorial office of this journal.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Errichiello E, Novara F, Cremante A, et al. Dissection of partial 21q monosomy in different phenotypes: clinical and molecular characterization of five cases and review of the literature. Mol Cytogenet 2016;9:21. [Crossref] [PubMed]
- Guion-Almeida ML, Richieri-Costa A, Jehee FS, et al. Frontonasal dysplasia, callosal agenesis, basal encephalocele, and eye anomalies syndrome with a partial 21q22.3 deletion. Am J Med Genet A 2012;158A:1676-9. [Crossref] [PubMed]
- Roberson ED, Wohler ES, Hoover-Fong JE, et al. Genomic analysis of partial 21q monosomies with variable phenotypes. Eur J Hum Genet 2011;19:235-8. [Crossref] [PubMed]
- Lyle R, Béna F, Gagos S, et al. Genotype-phenotype correlations in Down syndrome identified by array CGH in 30 cases of partial trisomy and partial monosomy chromosome 21. Eur J Hum Genet 2009;17:454-66. [Crossref] [PubMed]
- Wang L, Qu G, Tang X, et al. Child neglect and its association with social living ability: does the resilience attenuate the association? Psychol Health Med 2019;24:519-29. [Crossref] [PubMed]
- Egger G, Roetzer KM, Noor A, et al. Identification of risk genes for autism spectrum disorder through copy number variation analysis in Austrian families. Neurogenetics 2014;15:117-27. [Crossref] [PubMed]
- Sgardioli IC, Copelli MM, Lustosa-Mendes E, et al. Pure 21q22.3 deletion identified in a patient with mild phenotypic features. Congenit Anom (Kyoto) 2018;58:178-80. [Crossref] [PubMed]
- Chen CP, Lin SP, Chern SR, et al. De novo satellited 21q associated with corpus callosum dysgenesis, colpocephaly, a concealed penis, congenital heart defects, and developmental delay. Genet Couns 2004;15:437-42. [PubMed]
- Oegema R, de Klein A, Verkerk AJ, et al. Distinctive Phenotypic Abnormalities Associated with Submicroscopic 21q22 Deletion Including DYRK1A. Mol Syndromol 2010;1:113-20. [Crossref] [PubMed]
- Zeng J, Huang W, Huang M, et al. The first report showing de novo partial 21q monosomy in an adult woman with occult primary ovarian insufficiency (POI). Clin Chem Lab Med 2019;57:e230-3. [Crossref] [PubMed]
- Yao G, Chen XN, Flores-Sarnat L, et al. Deletion of chromosome 21 disturbs human brain morphogenesis. Genet Med 2006;8:1-7. [Crossref] [PubMed]
- Yamamoto T, Shimojima K, Nishizawa T, et al. Clinical manifestations of the deletion of Down syndrome critical region including DYRK1A and KCNJ6. Am J Med Genet A 2011;155A:113-9. [Crossref] [PubMed]
- Ruiz-Botero F, Pachajoa H. Deletion 21q22.3 and duplication 7q35q36.3 in a Colombian girl: a case report. J Med Case Rep 2016;10:204. [Crossref] [PubMed]
- Valetto A, Orsini A, Bertini V, et al. Molecular cytogenetic characterization of an interstitial deletion of chromosome 21 (21q22.13q22.3) in a patient with dysmorphic features, intellectual disability and severe generalized epilepsy. Eur J Med Genet 2012;55:362-6. [Crossref] [PubMed]
- Briegel W, Hoyer J. Psychiatric Disorders and Distal 21q Deletion-A Case Report. Int J Environ Res Public Health 2020;17:3096. [Crossref] [PubMed]
- Zhang Y, Tu B, Jiang X, et al. Exposure to carbon black nanoparticles during pregnancy persistently damages the cerebrovascular function in female mice. Toxicology 2019;422:44-52. [Crossref] [PubMed]
- Jasmin SB, Pearson V, Lalonde D, et al. Differential regulation of ABCA1 and ABCG1 gene expressions in the remodeling mouse hippocampus after entorhinal cortex lesion and liver-X receptor agonist treatment. Brain Res 2014;1562:39-51. [Crossref] [PubMed]
- Pereira PL, Magnol L, Sahún I, et al. A new mouse model for the trisomy of the Abcg1-U2af1 region reveals the complexity of the combinatorial genetic code of down syndrome. Hum Mol Genet 2009;18:4756-69. [Crossref] [PubMed]
- Tan TY, Sedmík J, Fitzgerald MP, et al. Bi-allelic ADARB1 Variants Associated with Microcephaly, Intellectual Disability, and Seizures. Am J Hum Genet 2020;106:467-83. [Crossref] [PubMed]
- Ganesh L, Yoshimoto T, Moorthy NC, et al. Protein methyltransferase 2 inhibits NF-kappaB function and promotes apoptosis. Mol Cell Biol 2006;26:3864-74. [Crossref] [PubMed]
- Baumann C, Olson M, Wang K, et al. Arginine methyltransferases mediate an epigenetic ovarian response to endometriosis. Reproduction 2015;150:297-310. [Crossref] [PubMed]
- Kong R, Shao S, Wang J, et al. Genetic variant in DIP2A gene is associated with developmental dyslexia in Chinese population. Am J Med Genet B Neuropsychiatr Genet 2016;171B:203-8. [Crossref] [PubMed]
- Hansen J, Snow C, Tuttle E, et al. De novo mutations in SIK1 cause a spectrum of developmental epilepsies. Am J Hum Genet 2015;96:682-90. [Crossref] [PubMed]
- Pröschel C, Hansen JN, Ali A, et al. Epilepsy-causing sequence variations in SIK1 disrupt synaptic activity response gene expression and affect neuronal morphology. Eur J Hum Genet 2017;25:216-21. [Crossref] [PubMed]



