
Identification and evaluation of fructose-bisphosphate aldolase B as a potential diagnostic biomarker in choledochal cysts patients: a quantitative proteomic analysis
Introduction
Choledochal cyst (CC) is a congenital bile duct malformation and is considered to be a precancerous state. CC occurs in patients of all ages, and is primarily diagnosed in children. The majority of CC is diagnosed before the age of 10, whereas only 20% are diagnosed in adulthood (1). Although the incidence of CC is approximately 1:100,000–1:150,000 in the western countries, it is about 1:1,000 in the Asian countries, especially in Japan (2,3). Moreover, with the development of diagnostic methods, more and more patients with CC are diagnosed (4) and there are three times more female patients than male patients (5). In patients with CC, the risks of associated complications such as intrahepatic stenosis and calculi, lobular liver atrophy/hypertrophy, secondary biliary cirrhosis, portal hypertension, liver failure, and biliary malignancies, are extremely high. CC is also associated with precancerous states that occur more frequently and earlier in these patients (2,5). The incidence of cholangiocarcinoma in patients with CC is between 9–28%. The histological types of CC observed in patients are as follows: adenocarcinoma (73–84%), anaplastic carcinoma (10%), undifferentiated cancer (5–7%), squamous cell carcinoma (5%), and other cancers (1.5%) (2,6). Common surgical methods such as external drainage of the common bile duct, internal drainage for dilated common bile duct and enteral anastomosis, common bile duct dilation, cholecystectomy, pancreaticobiliary shunt of common hepaticointestinal anastomosis, and bile duct reconstruction surgery, are considered radical surgery for common bile duct cysts. The treatment aims to avoid a large number of hepatic, pancreatic or biliary complications that may occur (7). However postoperative patients are still at risk of pancreatitis and ascending cholangitis (8). Since the early diagnosis of CC favors the treatment and prognosis of CC, it is still demanding in powerful manners for CC screen.
The clinical presentation of CC varies and is most often accompanied by nonspecific abdominal pain. In infants, it usually manifests as obstructive jaundice and abdominal masses. However, in adults, the classic triad of intermittent abdominal pain, jaundice, and a clear upper right abdominal mass occurs only in less than 20% of patients and the most common symptoms are pain, fever, nausea, vomiting, and jaundice, which are usually caused by complications related to ascending cholangitis and pancreatitis (9). The diagnosis of CC is typically based on transcorporeal ultrasound (US). If necessary, multimodality imaging techniques are also utilized, including computed tomography (CT), magnetic resonance imaging (MRI), and/or endoscopic retrograde cholangiopancreatography (ERCP), to determine the extent of ductal involvement (10). Although highly sensitive, ERCP is utilized less frequently due to its invasiveness and associated risks such as cholangitis, bleeding, pancreatitis, and perforation (11). Magnetic resonance cholangiopancreatography (MRCP) is a noninvasive diagnostic method for CC with high sensitivity and specificity (12-14). Furthermore, it can reliably diagnose CC-associated cholangiocarcinoma, choledocholithiasis and other complications. However, none of these imaging/diagnostic modalities, including MRCP, are suitable for routine screening of CC in high-risk populations due to the financial costs involved. Therefore, it still requires novel manners for CC early diagnosis. Many diseases can cause changes in the protein composition of serum. The discovery and detection of serum markers will facilitate the early screening and diagnosis of diseases (15). Therefore, a specific and sensitive biomarker for serology is of great value for CC diagnostics.
Serology is a minimally invasive technology and is especially useful in situations where biopsy specimens are not available (as in cases involving children or uncooperative adults). Some serological methods, such as the keratinocyte binding assays, may provide more information to confirm or rule out the diagnosis, especially in the absence of a biopsy (16). In many cases, combined with compatible clinical images, serological tests can be sufficient to confirm a diagnosis (17). Serum analysis relies on indirect immunofluorescence (IF) microscopy using natural tissue sections and recombinant proteins as substrates. Recombinant antigens are also used in western blot (WB), immunoprecipitation analysis or enzyme-linked immunosorbent assays (ELISA), and the latter has other relevance to the monitoring of disease activity. Traditionally, serological diagnosis follows a multi-step method based on the use of one or two tissue substrates for initial IF screening, followed by individual antigen-specific assays. At the same time, multi-parameter analysis tools can be used for efficient and rapid testing (18,19). This current study explored the differentially expressed proteins in paired CC and normal tissues using label-free liquid chromatography-mass spectrometry (LC-MS), with the aim of identifying reliable biomarkers for the diagnosis of CC. Among the differentially expressed proteins identified using label-free quantitative proteomic analysis, the expression profile of insulin-like growth factor binding protein 2 (IGFBP2), tropomyosin (TPM3), fructose-bisphosphate aldolase B (ALDOB), fumarylacetoacetate hydrolase (FAH), superoxide dismutase 3 (SOD3), secreted protein acidic and cysteine rich (SPARC), apolipoprotein E (APOE), and retinol binding protein 4 (RBP4) in the serum of CC patients were further examined using quantitative polymerase chain reactions (Q-PCRs), WB analyses, and immunohistochemistry (IHC) assays, supporting that ALDOB could serve as a potential effective biomarker for patients with CCs. We present the following article in accordance with the MDAR reporting checklist (available at https://dx.doi.org/10.21037/tp-21-336).
Methods
Collection of clinical samples
Patients diagnosed with type I CC by at least two specialists and who underwent surgical therapy at the Children’s Hospital of Soochow University from September 2018 to March 2020 were enrolled in this study. Three samples of hepatic tissues and serum were collected from each CC patients and healthy donors. Q-PCR, WB, and IHC were applied to analyze the samples. Patient clinical information is listed in Table S1. The collection and usage of tissue samples in this work was approved by the Ethics Committee for Human Research of the Children’s Hospital of Soochow University (No. 20160106011). Written informed consents were obtained from all participants. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Protein extraction
Serum samples were collected and clotted at room temperature, centrifuged at 1,000 ×g for 3 minutes at 4 °C and stored at −80 °C until further analysis. High-abundance proteins were removed with Pierce Top 12 (cat 85165, Thermo Scientific, Rockform, IL, USA) and the total protein concentration was measured using the bicinchonic acid assay kit (cat 23227, Pierce, Thermo Scientific, Rockform, IL, USA). Total protein (350 µg) was dried in a high-speed vacuum (Savant, Thermofisher) and dissolved in 6 M urea and 100 mM Tris-HCl (pH 7.4). Disulfide bonds were reduced with 10 mM dithiothreitol (cat D9760-1G, DTT, Sigma-Aldrich, St Louis, MO, USA) for 60 minutes at room temperature. The proteins were then alkylated with 30 mM iodoacetamide (cat I6125-5G, Sigma-Aldrich, St Louis, MO, USA) in the dark at room temperature for 60 minutes. The excess iodoacetamide was consumed by adding DTT (30 mM) and the sample was diluted 1:10 with high-purity Milli-Q water (Millipore, Billerica, MA, USA) before trypsin (Promega, Madison, Wisconsin) digestion at 37 °C for 18 hours. The sample was finally purified in Pierce C18 Spin Columns (Thermo Scientific, Rockform, IL, USA), dried, and dissolved in 0.1% formic acid (cat 00940, Sigma-Aldrich, St Louis, MO, USA) containing 12.5 M Hi3 peptide mixture (Waters).
Liquid chromatography-mass spectrometry analysis
The peptides were dissolved in the mobile phase A of liquid chromatography and separated using the EASY-nLC 1,000 ultra-high performance liquid system (Thermo Scientific, Rockform, IL, USA). Mobile phase A is an aqueous solution containing 0.1% formic acid and 2% acetonitrile, and mobile phase B is an aqueous solution containing 0.1% formic acid and 90% acetonitrile. The flow rate was maintained at 500.00 nl/min. The peptides were separated by the ultra-high performance liquid system and injected into the NSI ion source for ionization and analyzed by the QE+2 MS analyzer. The ion source voltage was set to 2.0 kV, and the peptide precursor ions and their secondary fragments were detected and analyzed by high-resolution Orbitrap (Thermo Scientific, Rockform, IL, USA). The scanning range of the primary MS was set to 350-1,800 m/z, and the scanning resolution was set to 70,000.00; the scanning range of the secondary MS was set to a fixed starting point of 100 m/z, and the secondary scanning resolution was set to 17,500.00. In the data acquisition mode, the data dependent scanning (DDA) program was used. The parent ion of the first 20.00 peptide segment with the highest signal strength was selected to enter the HCD collision pool in turn after the primary scanning, and the fragmentation energy was 28% for fragmentation, and the secondary MS was also conducted in turn. To improve the effective utilization of MS, automatic gain control (AGC) was set to 5E4, signal threshold was set to 4E4ions/s, the maximum injection time was set to 50 ms, and dynamic exclusion time of tandem mass scan was set to 30 seconds to avoid repeated scan of precursor ions. The peptides were quantified using label-free quantitative proteomics analysis, which is based on the measurement of mass spectrometric signal intensities of peptide precursor ions. Briefly, the corresponding signal abundance of the protein in each sample was detected by MS technology, and the LFQ intensity of the protein in each sample was obtained by non-standard quantitative calculation method. The ratio of protein LFQ intensity between two different samples was taken as the differential expression amount (ratio) of the comparison group.
Database search and quantification
Secondary mass spectrometry data were retrieved using Maxquant v.1.5.2.8. Retrieval parameters were set as follows: the database was Homo_sapiens_9606 (20366 sequences); an anti-library was added to calculate the false positive rate (FDR) caused by random matching; and a common contamination library was added to the database to eliminate the influence of contaminated proteins in the identification results. The restriction enzyme digestion method was set to Trypsin/P and the number of missed cleavage sites was set to 2. The minimum peptide length was set to 7 amino acid residues, the maximum modification number of peptides was set to 5, first search and main search primary precursor ion. The mass error tolerance was set to 20.0 and 5 ppm, respectively, and the mass error tolerance of the secondary fragment ion was 0.02 Da. Oxidation of cysteine Carbamidomethyl (C) was set to fixed modification and variable modification to [‘Acetyl (Protein N-term)’, ‘Oxidation (M)’, ‘Deamidation (NQ)’]. The quantitative method was set as LFQ, and the PROTEIN identification and PSM identification FDR were set as 1%.
Analysis of variance (ANOVA) was used to quantify significantly different protein expression levels (significance set at P<0.05). Principal component analysis (PCA) was used to visualize to the main axes of variation in different groups.
Gene Ontology (GO) analysis
GO annotations on proteomics were derived from the UniProt-GOA database (http://www.ebi.ac.uk/GOA/). First, the protein ID was converted to UniProt ID, then UniProt ID was used to match the GO ID, and the corresponding information from the UniProt-GOA database was retrieved according to GO ID. If there was no protein information in the UniProt-GOA database, an algorithm software based on protein sequence, InterProScan, was used to predict the GO function of the protein. The protein was then classified according to cellular components, molecular functions, or physiological processes.
Protein domain annotation
The software InterProScan (https://github.com/ebi-pf-team/interproscan) which is based on protein sequence algorithm, and the corresponding InterPro domain database were used to annotate the protein domains of the identified proteins. InterPro domain database (http://www.ebi.ac.uk/interpro/) is a free online database that integrates information including protein family classification, protein domain classification, and protein functional site classification. The core is based on the pattern or characteristics of the domains in the database. By evaluating the sequence of the submitted protein, the corresponding algorithm was used to determine the domain matched by the protein.
Subcellular localization
The WoLF PSORT program (https://psort.hgc.jp/) (20) was used to predicted protein subcellular localization.
GO enrichment analysis
Fisher’s exact test was used to assess the differentially expressed proteins in the context of the identified protein, and in the GO enrichment tests, a P value less than 0.05 was considered significant.
Quantitative polymerase chain reaction of the differential expressed protein genes
Q-PCR were performed on the selected eight protein targets in CC and normal choledochus tissues. Total RNA was extracted using TRIzol reagent (cat 15596018 Thermofisher Scientific, USA) following the manufacturer’s protocol. 1 µg of total RNA was reverse-transcribed into cDNA using PrimeScript miRNA cDNA Synthesis kit (cat D350A, Takara Bio, Inc.). Next, the cDNA was analyzed by Q-PCR system using SYBR green (SsoAdvanced Universal SYBR Green Supermix, cat. 1725274, Bio-Rad Laboratories, Inc). The gene expression level was calculated using the 2−ΔΔCT method and normalized by U6 or glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The primers used in this study are presented as follows: insulin-like growth factor-binding protein 2 (IGFBP2): forward 5'-GCAAGGGTGGCAAGCATC-3', reverse 5'-GCAAGGGTGGCAAGCATC-3'; retinol-binding protein 4 (RBP4): forward 5'-GAGCAAGATGGAATGGGTT-3', reverse 5'-CTCGGTGTCTGTAAAGGTG-3'; tropomyosin alpha-3 chain (TPM3): forward 5'-TCCTCTTACGGGGTGCTCTT-3', reverse 5'-GTTCCTGCCTTCCAGGTCAT-3'; fructose-bisphosphate aldolase B (ALDOB): forward 5'-TAGGATCCATGGCCCACCGATTTCCAGC-3', reverse 5'-TACTCGAGGTAGGTATAGCAGGCTGTGA-3'; fumarylacetoacetase (FAH): forward 5'-AGAATGCGCTGTTGCCAAA-3', reverse 5'-GGAAGCTCGGCCATGGTAT-3'; extracellular superoxide dismutase [Cu-Zn] (SOD3): forward 5'-TCGTCCTCTTCCGGCAGCTT-3', reverse 5'-GCTTCTTGCGCTCTGAGTGCT-3'; SPARC: forward 5'-GACCAACAGGGAAAACCTCA-3', reverse 5'-TGCAGGCTCCAAAATAATCC-3'; and apolipoprotein E (APOE): forward 5'-TCTGTGGCTACCAACTCCATTG-3', reverse 5'-GGCGTAGGTGAGGGATGATC-3'.
Western blotting assay
According to the results of proteome sequencing, WB was performed on the selected eight protein targets. Tissues were collected and homogenized using RIPA buffer (Beyotime Institute of Biotechnology). Equal amounts of total protein (30 µg/lane) were separated using 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE; Bio-Rad, USA). The primary antibodies were anti-IGFBP2 (ab227881, 1:1,000, Abcam, USA), anti-TPM3 (ab190993, 1:1,000, Abcam, USA), anti-ALDOB (ab129728, 1:1,000, Abcam, USA), anti-FAH (PA5-42049, 1:1,000, Invitrogen, USA), anti-SOD3 (14316-1-AP, 1:1,000, Proteintech, China), anti-SPARC (ab55847, 1:1,000, Abcam, USA), anti-APOE (ab52607, 1:1,000, Abcam, USA), anti-RBP4 (ab154914, 1:1,000, Abcam, USA), and anti-GAPDH (ab8245, 1:5,000, Abcam, USA). And the secondary antibody goat was anti-mouse IgG (PAB31797; Abnova, China). The bands were then developed using enhanced chemiluminescence chromogenic substrate (GE Healthcare, UK) and analyzed by the Image J software. GAPDH was used as control in this study.
Histopathology and immunohistochemical assay
Tissue samples were embedded in paraffin and cut into 5 µm slices. Sections were deparaffinized, rehydrated, boiled and dried. For hematoxylin and eosin (H&E) assay, the slides were stained with hematoxylin then counterstained with eosin, subsequently visualized under a light microscopy (Olympus Corporation) for histopathological examination.
For IHC assay, the slides were blocked with 1% bovine serum albumin (BSA) before incubation with an anti-ALDOB primary antibody (ab129728, 1:50, Abcam, USA) overnight at 4 °C. Slides were then incubated with the corresponding horseradish peroxidase (HRP)-conjugated secondary antibody (ab97051, 1:5,000, Abcam) at room temperature for 30 minutes. The slices were visualized using a 3,3 N-Diaminobenzidine Tertrahydrochloride (DAB) kit (Thermo Fisher Scientific, Inc.). The nuclei were counterstained with hematoxylin (Beyotime Institute of Biotechnology) according to the manufacturer’s protocol. Stained tissues were visualized using a light microscope under 20× magnification.
Statistical analysis
Statistical analysis was performed using the SPSS package (Windows version, v. 22, Chicago, IL, USA). Statistical significance between multiple experimental groups or two groups were assessed using ANOVA with Tukey’s multiple comparison tests or Student’s t-test (two-tailed), respectively. At least three independent replicants were carried out. P<0.05 denoted statistical significance.
Results
Differentially expressed proteins in serum samples from patients with CCs and healthy donors
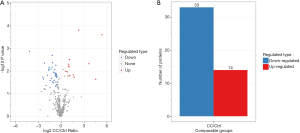
In this study, a total of 6,031 peptides were identified through spectral analysis, of which, 5,708 were specific peptides. A total of 670 proteins were identified, of which, 394 could be quantified. Among these proteins, 47 were differentially expressed in CC and normal serum samples, including 14 up-regulated and 33 down-regulated proteins (Figure 1).
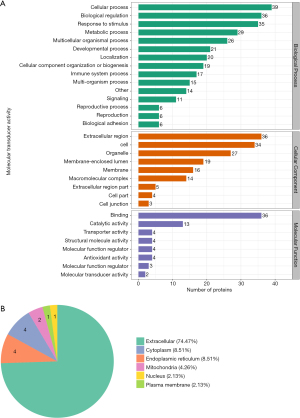
GO secondary annotation and subcellular structure location classification
GO classification revealed the biological functions of the differentially expressed proteins identified in the serum. The differentially expressed proteins are mostly involved in cellular processes, biological regulation, stimulus responses, metabolic processes, and multicellular biological processes. Most of the differentially expressed proteins can be localized to extracellular regions, cells, organelles, membrane enclosed cavities, and membranes. However, in terms of molecular functions these differentially expressed proteins are largely involved in binding and catalytic activities (Figure 2A). Moreover, the subcellular structure location was consistent with the above GO secondary classification results which showed that most of the differentially expressed proteins were localized outside the cell (Figure 2B).
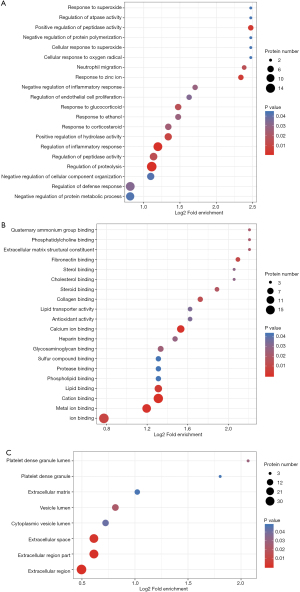
GO enrichment analysis
Differential protein enrichment analyses were carried out (Figure 3). The results showed that the differentially expressed proteins were enriched in the biological processes of proteolysis regulation and inflammatory response, and in molecular functions involving ion, metal ion, and cation binding processes. The differentially expressed proteins were enriched outside the cell.
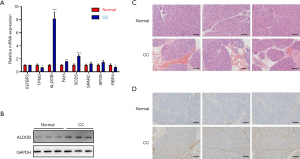
Histopathology and immunohistochemical (IHC) verification analysis
According to the differential expression profile and the references (Table S2), eight differentially expressed proteins, including insulin-like growth factor-binding protein 2 (IGFBP2), retinol-binding protein 4 (RBP4), tropomyosin alpha-3 chain (TPM3), fructose-bisphosphate aldolase B (ALDOB), fumarylacetoacetase (FAH), extracellular superoxide dismutase [Cu-Zn] (SOD3), SPARC, and apolipoprotein E (APOE), were selected for further investigation. Both the mRNA and protein levels of ALDOB were significantly higher in hepatic tissues from CC patients compared to healthy controls (Figure 4A,B). H&E staining and IHC assays demonstrated that hepatic tissues from CC patients demonstrated epithelial hyperplasia with round cell infiltration, bile duct wall thickening, and fibrosis (Figure 4C), as well as high expression of ALDOB (Figure 4D).
Discussion
CC is characterized by single or multiple cystic dilation of the intrahepatic and/or extrahepatic bile ducts. Disease-specific complications include cholangitis, portal hypertension, and malignant tumors (21). Additionally, incidence of CC is higher in Asian populations (about 1 in 1,000) compared to Western populations (1 in 100,000) (3,22). Although CC is mainly diagnosed in children, the incidence in the adult population has gradually increased (7).
Surgery, specifically cyst resection and hepatojejunostomy, are the treatments of choice for patients with CC (23). Hepatoduodenal anastomosis may be performed for biliary reconstruction. Postsurgical complications of CC include anastomotic stenosis or leakage, bleeding, small bowel obstruction, and internal hernia (21). Decades after the initial excision of CC, malignant tumors of the residual ducts can still present as a cause for concern. A retrospective study of Japanese patients with CC showed that the incidence of cholangiocarcinoma was 16.2% (24). These data strongly indicated that CC requires early diagnosis and long-term monitoring. Currently, transcorporeal ultrasound (US) and multimodality imaging techniques are the preferred diagnostic methods for CC due to the advantages of non-invasive, repeatable inspection and dynamic observation. They can display the choledochal cysts intuitively, sensitively and accurately. However, they are still “technology” dependent method, which rely on the skills and experience of the operators, and take a long time. Serum marker examination, as a non-invasive and rapid detection method, will be an effective auxiliary method of ultrasound examination in CC examination, which could even be conducted even at the bedside. Therefore, development of an effective biomarkers for early screening and diagnosis is crucial in CC therapy and patient prognosis.
This work was designed to identify potential biomarkers in serum samples from CC patients. Quantitative label free proteomic analysis was used to identify differentially expressed proteins between CC and normal tissues. A total of 670 proteins were identified, of which, 394 could be quantified. The expression profile of eight biomarker candidates, namely, IGFBP2, TPM3, ALDOB, FAH, SOD3, SPARC, APOE, and RBP4, were further examined using Q-PCR, WB, and immunohistochemistry. The results demonstrated that the ALDOB protein highly expressed in both choledochal tissues and serum samples of CC patients, and may act as an effective potential biomarker for patients with CC.
ALDOB is expressed in many human tissues, especially in the liver (25,26). Down-regulation of ALDOB is considered an indicator for aggressive tumor characteristics, such as low differentiation, high TNM staging, early tumor recurrence, and short 5-year survival rate (27). ALDOB expression has been shown to be significantly reduced in gastric cancer and clear cell renal cell carcinoma tissues compared to adjacent normal tissues (26,28). In fact, gastric cancer patients showing low ALDOB expression had significantly reduced long-term survival compared to patients with high ALDOB expression (29). Another study demonstrated that stable expression of ALDOB inhibited liver metastasis, suggesting that low expression of ALDOB was associated with poor prognosis (30,31). These studies all suggest that ALDOB may be a potential prognostic biomarker and therapeutic target for a range of malignancies. On the contrary, some studies have suggested that high levels of ALDOB may be associated with certain cancers or diseases. Silencing the expression of ALDOB inhibited the proliferation and invasion of colorectal cancer cells by increasing the epithelial marker E-cadherin, and decreasing the mesenchymal markers N-cadherin and vientin (32). This suppression is absent in normal cells expressing ALDOB (33). The liver microenvironment can cause liver metastasis of colorectal cancer in mice and up-regulate ALDOB expression. ALDOB is responsible for the cleavage of fructose 1-phosphate into dihydroxyacetone phosphate and glyceraldehyde (34,35), which is believed to be associated with liver metastases. In addition, elevated plasma ALDOB levels were observed in the mouse non-alcoholic fatty liver model and excessive intake of fructose was the main driving force (36). The increased ALDOB level may result from cell leakage due to inflammation of the liver cells caused by excessive fat accumulation (37). This current work confirmed an obvious up-regulation of ALDOB in choledochal tissues, which may play an important role in the progression of CC, however further studies investigating the exact molecular pathways and mechanisms of ALDOB in CC are warranted. Moreover, the significantly increased expression level of ALDOB in the serum sample of CC patients may be a potential novel biomarker for early diagnostic biomarkers for CC.
Acknowledgments
Funding: None.
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://dx.doi.org/10.21037/tp-21-336
Data Sharing Statement: Available at https://dx.doi.org/10.21037/tp-21-336
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tp-21-336). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All experiments were approved by the Ethics Committee for Human Research of the Children’s Hospital of Soochow University (No. 20160106011), and written informed consent was obtained from all patients. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Atkinson HD, Fischer CP, de Jong CH, et al. Choledochal cysts in adults and their complications. HPB (Oxford) 2003;5:105-10. [Crossref] [PubMed]
- Moslim MA, Takahashi H, Seifarth FG, et al. Choledochal Cyst Disease in a Western Center: A 30-Year Experience. J Gastrointest Surg 2016;20:1453-63. [Crossref] [PubMed]
- Madadi-Sanjani O, Petersen C. Choledochal Cyst. In: Lacher M, St. Peter SD, Zani A. editors. Pearls and Tricks in Pediatric Surgery. Cham: Springer; 2021: 265-70.
- Dhupar R, Gulack B, Geller DA, et al. The changing presentation of choledochal cyst disease: an incidental diagnosis. HPB Surg 2009;2009:103739 [Crossref] [PubMed]
- Kabra V, Agarwal M, Adukai TK, et al. Choledochal cyst: a changing pattern of presentation. ANZ J Surg 2001;71:159-61. [Crossref] [PubMed]
- O'Neill JA Jr. Choledochal cyst. Curr Probl Surg 1992;29:361-410. [Crossref] [PubMed]
- Ronnekleiv-Kelly SM, Soares KC, Ejaz A, et al. Management of choledochal cysts. Curr Opin Gastroenterol 2016;32:225-31. [PubMed]
- Franko J, Nussbaum ML, Morris JB. Choledochal cyst cholangiocarcinoma arising from adenoma: case report and a review of the literature. Curr Surg 2006;63:281-4. [Crossref] [PubMed]
- Singham J, Yoshida EM, Scudamore CH. Choledochal cysts: part 2 of 3: Diagnosis. Can J Surg 2009;52:506-11. [PubMed]
- Martin RF. Biliary cysts: a review and simplified classification scheme. Surg Clin North Am 2014;94:219-32. [Crossref] [PubMed]
- Saito T, Terui K, Mitsunaga T, et al. Role of pediatric endoscopic retrograde cholangiopancreatography in an era stressing less-invasive imaging modalities. J Pediatr Gastroenterol Nutr 2014;59:204-9. [Crossref] [PubMed]
- Park DH, Kim MH, Lee SK, et al. Can MRCP replace the diagnostic role of ERCP for patients with choledochal cysts? Gastrointest Endosc 2005;62:360-6. [Crossref] [PubMed]
- Huang CT, Lee HC, Chen WT, et al. Usefulness of magnetic resonance cholangiopancreatography in pancreatobiliary abnormalities in pediatric patients. Pediatr Neonatol 2011;52:332-6. [Crossref] [PubMed]
- Sacher VY, Davis JS, Sleeman D, et al. Role of magnetic resonance cholangiopancreatography in diagnosing choledochal cysts: Case series and review. World J Radiol 2013;5:304-12. [Crossref] [PubMed]
- Nossov V, Amneus M, Su F, et al. The early detection of ovarian cancer: from traditional methods to proteomics. Can we really do better than serum CA-125? Am J Obstet Gynecol 2008;199:215-23. [Crossref] [PubMed]
- Saschenbrecker S, Karl I, Komorowski L, et al. Serological Diagnosis of Autoimmune Bullous Skin Diseases. Front Immunol 2019;10:1974. [Crossref] [PubMed]
- Schmidt E, Zillikens D. The diagnosis and treatment of autoimmune blistering skin diseases. Dtsch Arztebl Int 2011;108:399-405. I-III. [Crossref] [PubMed]
- van Beek N, Rentzsch K, Probst C, et al. Serological diagnosis of autoimmune bullous skin diseases: prospective comparison of the BIOCHIP mosaic-based indirect immunofluorescence technique with the conventional multi-step single test strategy. Orphanet J Rare Dis 2012;7:49. [Crossref] [PubMed]
- van Beek N, Dähnrich C, Johannsen N, et al. Prospective studies on the routine use of a novel multivariant enzyme-linked immunosorbent assay for the diagnosis of autoimmune bullous diseases. J Am Acad Dermatol 2017;76:889-894.e5. [Crossref] [PubMed]
- Horton P, Park KJ, Obayashi T, et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res 2007;35:W585-7 [Crossref] [PubMed]
- Ono S, Sakai K, Kimura O, et al. Development of bile duct cancer in a 26-year-old man after resection of infantile choledochal cyst. J Pediatr Surg 2008;43:E17-9. [Crossref] [PubMed]
- Soares KC, Arnaoutakis DJ, Kamel I, et al. Choledochal cysts: presentation, clinical differentiation, and management. J Am Coll Surg 2014;219:1167-80. [Crossref] [PubMed]
- Forny DN, Ferrante SM, Silveira VG, et al. Choledochal cyst in childhood: review of 30 cases. Rev Col Bras Cir 2014;41:331-5. [Crossref] [PubMed]
- Watanabe Y, Toki A, Todani T. Bile duct cancer developed after cyst excision for choledochal cyst. J Hepatobiliary Pancreat Surg 1999;6:207-12. [Crossref] [PubMed]
- Ji S, Zhang B, Liu J, et al. ALDOA functions as an oncogene in the highly metastatic pancreatic cancer. Cancer Lett 2016;374:127-35. [Crossref] [PubMed]
- Saito H. The distribution of the aldolase isoenzymes in various human tissues and the anomaly in cancerous tissues -especially in gastric cancer- (author's transl). Hokkaido Igaku Zasshi 1975;50:540-8. [PubMed]
- Peng SY, Lai PL, Pan HW, et al. Aberrant expression of the glycolytic enzymes aldolase B and type II hexokinase in hepatocellular carcinoma are predictive markers for advanced stage, early recurrence and poor prognosis. Oncol Rep 2008;19:1045-53. [Crossref] [PubMed]
- Asaka M, Kimura T, Meguro T, et al. Alteration of aldolase isozymes in serum and tissues of patients with cancer and other diseases. J Clin Lab Anal 1994;8:144-8. [Crossref] [PubMed]
- He J, Jin Y, Chen Y, et al. Downregulation of ALDOB is associated with poor prognosis of patients with gastric cancer. Onco Targets Ther 2016;9:6099-109. [Crossref] [PubMed]
- Tao QF, Yuan SX, Yang F, et al. Aldolase B inhibits metastasis through Ten-Eleven Translocation 1 and serves as a prognostic biomarker in hepatocellular carcinoma. Mol Cancer 2015;14:170. [Crossref] [PubMed]
- Ito S, Shen L, Dai Q, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011;333:1300-3. [Crossref] [PubMed]
- Li Q, Li Y, Xu J, et al. Aldolase B Overexpression is Associated with Poor Prognosis and Promotes Tumor Progression by Epithelial-Mesenchymal Transition in Colorectal Adenocarcinoma. Cell Physiol Biochem 2017;42:397-406. [Crossref] [PubMed]
- Bu P, Chen KY, Xiang K, et al. Aldolase B-Mediated Fructose Metabolism Drives Metabolic Reprogramming of Colon Cancer Liver Metastasis. Cell Metab 2018;27:1249-1262.e4. [Crossref] [PubMed]
- Malay AD, Allen KN, Tolan DR. Structure of the thermolabile mutant aldolase B, A149P: molecular basis of hereditary fructose intolerance. J Mol Biol 2005;347:135-44. [Crossref] [PubMed]
- Berardini TZ, Amsden AB, Penhoet EE, et al. Identification of conserved promoter elements for aldB and isozyme specific residues in aldolase B. Comp Biochem Physiol B Biochem Mol Biol 1999;122:53-61. [Crossref] [PubMed]
- DiNicolantonio JJ, Subramonian AM, O'Keefe JH. Added fructose as a principal driver of non-alcoholic fatty liver disease: a public health crisis. Open Heart 2017;4:e000631 [Crossref] [PubMed]
- Niu L, Geyer PE, Wewer Albrechtsen NJ, et al. Plasma proteome profiling discovers novel proteins associated with non-alcoholic fatty liver disease. Mol Syst Biol 2019;15:e8793 [Crossref] [PubMed]
(English Language Editor: J. Teoh)


