
A novel endoglin mutation in a family with hereditary hemorrhagic telangiectasia: a case report
Introduction
Hereditary hemorrhagic telangiectasia (HHT), also known as Osler-Weber-Rendu disease, is a rare autosomal dominant vascular disease with an annual incidence rate of 1:5,000 to 10,000 (1). It is characterized by mucosal telangiectasia, epistaxis, and visceral arteriovenous malformations (AVMs) predominantly in the lungs, liver, and brain (2). These AVMs lead to serious symptoms, such as hemoptysis, liver cirrhosis, cerebral hemorrhage, brain abscesses, pulmonary arterial hypertension (PAH), deep vein thrombosis, etc. (3,4). HHT diagnoses are currently based on the Curaçao criteria (1) as follows: (I) recurrent spontaneous epistaxis; (II) telangiectasia of the skin or mucosa of the lips, nose, mouth, fingers, etc.; (III) a family history of HHT (at least one first-degree relative with the condition); and (IV) visceral injury (gastrointestinal telangiectasia with or without bleeding, AVMs in the lungs, liver, brain, spleen, etc.). When three of these four criteria are satisfied, HHT can definitively be diagnosed. If two of the above criteria are met, a diagnosis of HHT is suspected. If only one of the above criteria is met, HHT is generally not diagnosed.
While obvious signs of HHT, such as telangiectasia of the face, mouth, or hands, are reported in younger patients aged 5–30 years, predominantly following the onset of nosebleeds, other manifestations, such as AVMs, are more difficult to identify in this age group (5). There is therefore a risk of missing an HHT diagnosis in children and young adults; the clinical diagnostic features of HHT differ between age groups, and the age of onset can vary. Prompted by the frequency of misdiagnosis in children and young adults, a multicenter study evaluated the use of the Curaçao criteria in the diagnosis of HHT patients between the ages of 0 and 21. This study found that gene detection is the method of choice to diagnose HHT in patients that met one or two Curaçao criteria (6).
As an inherited vascular disease, HHT is divided into three types: HHT1, involving endoglin (ENG) gene mutations; HHT2, involving ACVRL1 mutations; and HHT3, involving SMAD4 mutations (7). Approximately 80% of all HHT patients have mutations in the ENG or ACVRL1 genes (8). Pulmonary arteriovenous malformations (PAVMs) and cerebral arteriovenous malformations (CAVMs) occur most frequently in HHT1 (9).
In this case report, we discuss a 9-year-old girl with a heterozygous ENG gene mutation [c.1466del (p.Gln489Argfs*2)] that was first identified using whole-exome sequencing (WES) and confirmed by Sanger sequencing. The patient’s mother had a history of recurrent epistaxis and was also a carrier of the ENG mutation. The father was not a carrier. No mutations were found in other HHT-related genes. Using STRUM software, we demonstrated that this mutation affects the configuration of ENG protein. We present the following article/case in accordance with the CARE reporting checklist (available at https://dx.doi.org/10.21037/tp-20-156).
Case presentation
In September 2016, a 7-year-old girl experiencing tic symptoms and 4 days of fever was transferred to our hospital (Shanghai Children’s Hospital) from a local hospital due to intracranial bleeding. Brain computed tomography (CT), magnetic resonance imaging (MRI), and magnetic resonance angiography (MRA) performed at this local hospital indicated intracranial hemorrhage and CAVMs. Before her transfer to our hospital, the patient received emergency treatment that included mannitol, hemostasis, antiepileptic medication, neurotrophic medication, etc. At the time of her transfer, she still had a fever, but was no longer experiencing tic symptoms. Using brain CT and craniocerebral angiography, clinicians at our hospital confirmed that the cause of the patient’s intracranial bleeding was CAVM (Figure 1A,B). On the 2nd and 9th days following admission, the patient underwent brain craniocerebral angiography to locate the source of bleeding and cerebral vascular embolization procedures to stem its source. After the patient’s first operation, the presence of multiple CAVMs was confirmed and the source of bleeding was partially embolized. With a range of comprehensive treatments, including anti-infection, nutritional nerve, and anti-epilepsy drugs, the patient’s vital signs stabilized and she no longer experienced tic symptoms.
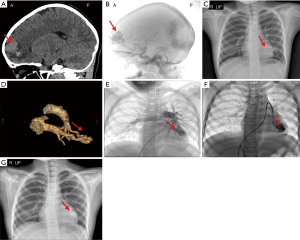
Prior to her third admission to hospital, the patient was experiencing headaches and an occasional cough. As a chest radiograph taken previously indicated enlarged vessels in the left lower lung (Figure 1C) and the patient’s chest CT suggested she may have a PAVM, we performed cardiac computed tomography angiography (CTA) and confirmed the presence of a PAVM in a segment of the left lung (S5) (Figure 1D). The patient’s abdominal CT enhancement was normal, and she was diagnosed as having CAVMs and PAVMs. As she had multiple CAVMs, the neurosurgeon suggested that cerebral vascular embolization therapy be performed in stages, and the patient attended the neurology department of our hospital between September 2016 and January 2018 to receive this therapy. Throughout this period, the patient experienced intracranial hemorrhage again, this time in the right temporal-occipital lobe. The patient did not display any clinical symptoms, and we administered the same treatment. During her final admission to the neurology department, the patient’s intracranial vascular condition was evaluated by craniocerebral angiography and brain CT and found to be stable. She did not require further embolization therapy and was warned against participating in strenuous exercise.
In August 2018, the patient, now 9 years old, attended the cardiology department at our hospital for PAVM treatment. The patient reported a history of untreated recurrent epistaxis for 2 years. Her mother also had a history of recurrent epistaxis. A vascular murmur (bruit) was detected in her precordium. The oxygen saturation (SpO2) of the left and right upper limbs was detected as 97% and 99%, respectively, on room air. Blood tests revealed a hemoglobin level of 14.6 g/dL. The patient’s electrocardiogram (ECG) and echocardiogram were normal. Her PAVMs were characterized as simple PAVMs with a feeding artery 6.7 mm in diameter as determined by angiography (Figure 1E).
As the patient had CAVMs, PAVMs, and a history of recurrent epistaxis, she met two of the four Curaçao criteria. We therefore proceeded with genetic testing to confirm a diagnosis of HHT. Once the patient’s condition improved, we elected to treat her PAVMs with transcatheter embolization therapy using a patent ductus arteriosus closure device (Figure 1F,G). Surgery was successful and the patient’s vital signs were stable; she experienced no chest tightness, hemoptysis, or other discomfort postsurgery. Her chest radiograph following surgery showed that the location of the closure device was correct. A month later, the patient again attended the cardiology department at our hospital for follow-up. Her outcomes were good, including her clinical symptoms, chest radiograph, ECG, and echocardiogram. Following this, the patient was followed up at her local hospital and has had no complaints to date (Figure 2).
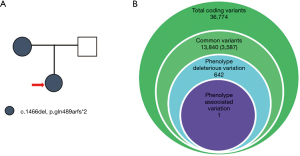
The process of genetic testing was as follows. A DNA library and WES were constructed according to the instructions of the manufacturer. Peripheral blood was collected from the patient and her family and genomic DNA was extracted using a DNA extraction kit (Tiangen, China) (Figure 3A). Following this, Exon trapping, gene amplification, and library building were performed using the Roche Nimblegen SeqCap EZ Exome V3 kit (Roche, Switzerland). Hiseq 2500 was used for sequencing. The average sample sequencing depth was 100×, and the sequencing depth of the core target sequence region was no less than 20×. Following the manufacturer’s instructions, whole genome sequencing were performed using Hiseq X Ten (Illumina, San Diego, CA, USA). The paired-end readings were then matched with the GRCh37/hg19 human reference sequence. As shown in Figure 3B, BAM and VCF files were analyzed using GATK and Clinvar software, and a total of 36,774 gene variants were detected. Common variants were filtered based on their minor allele frequencies (MAFs) (MAF <0.05) in the following databases: the Exome Aggregation Consortium (ExAC; http://exac.broadinstitute.org), Exome Sequencing Project (ESP; https://esp.gs.washington), 1000 Genomes (http://www.1000genomes.org), and the Genome Aggregation Database (gnomAD; http://gnomad.broadinstitute.org). By thoroughly analyzing all genes of the presumed dominant and recessive pathogenic variants, 13,840 variants were obtained, and 3,587 were further confirmed by filtering synonymous mutations, reserved base substitution mutations, shift mutations, non-shift mutations, termination mutations, link reading mutations, and splice mutations. Predictions made by SIFT, Polyphen2, Mutation Taster, Mutation Assessor, and annotations of the Human Gene Mutation Database (HGMD) and the Clinvar database were used to identify 642 potential pathogenic rare mutation sites. Finally, a mutation of the ENG gene was identified according to the phenotype of the patients with HHT syndrome. All such rare phenotypic variations are classified according to the American Society for Medical Genetics and Genomics/Molecular Pathology (ACMG/AMP) guidelines.
This identified deletion mutation of the ENG gene was verified by Sanger sequencing. In brief, primers were designed to partially amplify the region of the ENG gene that covered the mutation (ENST00000344849). The forward primer sequence was 5'-CAGCAGTGTCTTCCTGCATC-3', and the reverse primer sequence was 5'- GGCTGGAATTGTAGGCCAAG-3'. To further verify this gene mutation, polymerase chain reaction (PCR) products were isolated on agarose gel, purified with a QIAquick PCR purification kit (Qiagen, USA), and sent to Hong Zhong Biotechnology Co., Ltd. (Suzhou, China) for first-generation sequencing.
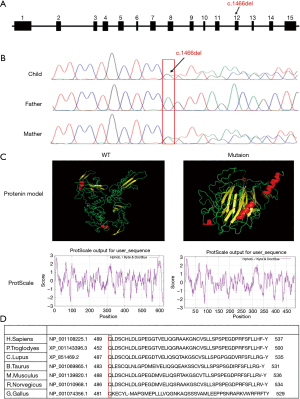
A heterozygous c.1466del (p.Gln489Argfs*2) mutation in the ENG gene was identified in this HHT pedigree by WES and confirmed by Sanger sequencing (Figure 4A,B). STRUM software was then used to predict the effect of a Gln489Argfs*2 site mutation on the stability of the ENG protein. This confirmed that the conformation of the ENG protein had changed: its hydrophilicity increased and its hydrophobicity decreased (Figure 4C). Importantly, the c.1466del mutation is located within a conserved region (Figure 4D), suggesting that this mutation may affect the function of the ENG protein.
All procedures involving human participants were performed in accordance with both the ethical standards of the Institutional Review Board of Shanghai Children’s Hospital and the Helsinki Declaration, as revised in 2013. Written informed consent was obtained from the patient and her parents.
Discussion
This case report yielded two principal findings: (I) the first account in China of an ENG p.Gln489Argfs*2 frameshift mutation, which was potentially a pathogenic gene mutation in the family of the case study patient; (II) a protein modeling analysis indicating that a c.1466del (p.Gln489Argfs*2) heterozygous mutation reduced the functioning of ENG proteins.
HHT is an inherited vascular disease in which symptoms develop over time (10). Compared with adults, children with HHT are less likely to show symptoms of the disease. Diagnosing children and teenagers with HHT is therefore very difficult (11). Genetic testing is therefore of particular importance. Previous studies have found that a greater prevalence of PAVMs and CAVMs occur with HHT1, and these PAVMs and CAVMs may be a result of ENG gene mutations (12). ENG mutations include deletions, insertions, copies, and nonsense mutations; these have been reported in more than 500 patients with HHT (http://arup.utah.edu/database/ENG/ENG_display.php). Nishida et al. (13) retrospectively analyzed 170 patients with HHT and CAVMs and found that mutations of the ENG gene were found in 75 of these patients (69%), and included c.1465_1466delCA (p.gln489fs), c.277C>T (p.arg93*), c.923C>A (p.la308asp), etc. The patient in the current case had a history of recurrent epistaxis, CAVMs, and PAVMs, and had undergone gene testing that indicated a mutation in her ENG gene. This information, combined with her mother’s similar history of recurrent epistaxis, led to the patient’s diagnosis of HHT1.
ENG is a type I transmembrane glycoprotein expressed in activated endothelial cells. This protein functions as an endothelial co-receptor for multiple receptor complexes of the TGF-β superfamily, which plays an important role in vascular remodeling (14). ENG comprises a short cytosolic tail, a single hydrophobic transmembrane domain, and a large extracellular region which contains an N-terminal orphan region (OR) and a C-terminal bipartite zona pellucida (ZP) module (15,16). Ruiz-Llorente et al. (17) found that a novel mutation in exon 9 of the ENG gene (c.1145 G>A), which involves Cys382 residue of ENG (p.Cys382>Tyr) in its ZP module, downregulates ENG expression, and thus has a pathogenic role in this variant of HHT1. ENG is expressed as a homodimer linked by intermolecular disulfide bonds. However, mutated ENG proteins may be misfolded and unstable. Due to this, they cannot reach the cell surface, rendering them incapable of forming heterodimers with normal ENG (7). Previous research has found that some ENG gene mutations, including c.1151T>C, c.1171G>T, c.1193A>G, c.1235G>A, c.1478G>A, and c.1585C>T, in the ZP module cause ENG protein to mutate. Mutant ENG can form heterodimers with wild type ENG, but this complex cannot reach the cell surface, reducing their function in vascular physiology (18). In our study, we found that a family pedigree of HHT with PAVMs and CAVMs was associated with a novel ENG gene mutation, c.1466del, which is located on the 12th exon of the ENG gene and causes p.Gln489Argfs*2 mutations in the ZP module of ENG. Large numbers of mutant ENG proteins with affected ZP modules interfere with hydrophobic core packing, which explains the impaired expression and/or secretion of the corresponding mutants (17). STRUM software predicted that ENG proteins with p.Gln489Argfs*2 mutations would have increased hydrophilicity and decreased hydrophobicity. Despite configuration changes of ENG with p.Gln489Argfs*2 mutations, these proteins can still form heterodimers with wild type ENG. However, these complexes cannot reach the cell surface. ENG gene mutations that result in ENG protein haploinsufficiency are thought to impair the recruitment of mural cells to vessels (19), which induces dysangiogenesis and AVMs.
We can only speculate from this case report what function ENG mutant proteins play in AVMs. However, by transfecting this mutation into cell lines and mouse models, future studies may be able to predict the function of these ENG mutant proteins.
In conclusion, we found that the newly identified c.1466del (p.Gln489Argfs*2) frameshift mutation was likely the molecular basis of disease in this HHT family. Genetic testing is therefore not only useful in diagnosing patients that demonstrate only one or two clinical characteristics of HHT, but can also assist in determining the molecular basis of the disease. Once this molecular basis is established and verified, further targeted treatments for HHT can be explored.
Acknowledgments
Funding: This work is supported by Research Fund for Shanghai Science and Technology Committee (18411965800, 19411963600), the Interdisciplinary Program of Shanghai Jiao Tong University (ZH2018QNA31, YG2016ZD05, ZH2018ZDA26) and National Natural Science Foundation of China (NSFC) (No. 81900437).
Footnote
Reporting Checklist: The authors have completed the CARE reporting checklist. Available at https://dx.doi.org/10.21037/tp-20-156
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/tp-20-156). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in studies involving human participants were in accordance with the ethical standards of the Institutional Review Board of Shanghai Children’s Hospital and with the Helsinki Declaration (as revised in 2013). Written informed consent was obtained from the patient and her parents.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Lacombe P, Lacout A, Marcy PY, et al. Diagnosis and treatment of pulmonary arteriovenous malformations in hereditary hemorrhagic telangiectasia: An overview. Diagn Interv Imaging 2013;94:835-48. [Crossref] [PubMed]
- Cartin-Ceba R, Swanson KL, Krowka MJ. Pulmonary arteriovenous malformations. Chest 2013;144:1033-44. [Crossref] [PubMed]
- Shovlin CL, Letarte M. Hereditary haemorrhagic telangiectasia and pulmonary arteriovenous malformations: issues in clinical management and review of pathogenic mechanisms. Thorax 1999;54:714-29. [Crossref] [PubMed]
- Khurshid I, Downie GH. Pulmonary arteriovenous malformation. Postgrad Med J 2002;78:191-7. [Crossref] [PubMed]
- Giordano P, Nigro A, Del Vecchio GC, et al. HHT in childhood: screening for special patients. Curr Pharm Des 2006;12:1221-5. [Crossref] [PubMed]
- Pahl KS, Choudhury A, Wusik K, et al. Applicability of the Curaçao Criteria for the Diagnosis of Hereditary Hemorrhagic Telangiectasia in the Pediatric Population. J Pediatr 2018;197:207-13. [Crossref] [PubMed]
- Abdalla SA, Letarte M. Hereditary haemorrhagic telangiectasia: current views on genetics and mechanisms of disease. J Med Genet 2006;43:97-110. [Crossref] [PubMed]
- McDonald J, Bayrak-Toydemir P, Pyeritz RE. Hereditary hemorrhagic telangiectasia: an overview of diagnosis, management, and pathogenesis. Genet Med 2011;13:607-16. [Crossref] [PubMed]
- Letteboer TG, Mager JJ, Snijder RJ, et al. Genotype-phenotype relationship in hereditary haemorrhagic telangiectasia. J Med Genet 2006;43:371-7. [Crossref] [PubMed]
- Govani FS, Shovlin CL. Hereditary haemorrhagic telangiectasia: a clinical and scientific review. Eur J Hum Genet 2009;17:860-71. [Crossref] [PubMed]
- Mowers KL, Sekarski L, White AJ, et al. Pulmonary arteriovenous malformations in children with hereditary hemorrhagic telangiectasia: a longitudinal study. Pulm Circ 2018;8:2045894018786696 [Crossref] [PubMed]
- Berg J, Porteous M, Reinhardt D, et al. Hereditary haemorrhagic telangiectasia: a questionnaire based study to delineate the different phenotypes caused by endoglin and ALK1 mutations. J Med Genet 2003;40:585-90. [Crossref] [PubMed]
- Nishida T, Faughnan ME, Krings T, et al. Brain arteriovenous malformations associated with hereditary hemorrhagic telangiectasia: gene-phenotype correlations. Am J Med Genet A 2012;158A:2829-34. [Crossref] [PubMed]
- Redgrave RE, Tual-Chalot S, Davison BJ, et al. Cardiosphere-derived cells require endoglin for paracrine-mediated angiogenesis. Stem Cell Reports 2017;8:1287-98. [Crossref] [PubMed]
- Bernabeu C, Conley BA, Vary CP. Novel biochemical pathways of endoglin in vascular cell physiology. J Cell Biochem 2007;102:1375-88. [Crossref] [PubMed]
- Saito T, Bokhove M, Croci R, et al. Structural basis of the human endoglin-BMP9 interaction: insights into BMP signaling and HHT1. Cell Rep 2017;19:1917-28. [Crossref] [PubMed]
- Ruiz-Llorente L, Chiapparino E, Plumitallo S, et al. Characterization of a mutation in the zona pellucida module of Endoglin that causes Hereditary Hemorrhagic Telangiectasia. Gene 2019;696:33-9. [Crossref] [PubMed]
- Förg T, Hafner M, Lux A. Investigation of endoglin wild-type and missense mutant protein heterodimerisation using fluorescence microscopy based IF, BiFC and FRET analyses. PLoS One 2014;9:e102998 [Crossref] [PubMed]
- Shovlin CL. Hereditary haemorrhagic telangiectasia: pathophysiology, diagnosis and treatment. Blood Rev 2010;24:203-19. [Crossref] [PubMed]
(English Language Editors: B. Madden and J. Gray)



