
Analysis of gut microbiota alteration and application as an auxiliary prognostic marker for sepsis in children: a pilot study
Introduction
Sepsis, defined as life-threatening organ dysfunction due to a dysregulated host response to infection, is a serious healthcare concern that affects neonatal, pediatric, and adult populations worldwide (1). Despite improvement in clinical care, pediatric sepsis is still a leading cause of morbidity, mortality and health care costs in infants and children (2). The reported global burden and mortality of neonatal and pediatric sepsis is grim, with an estimate of 48 cases and 22 severe cases in children per 100,000 person-year, and mortality of 11–19% (3). Early diagnosis, appropriate classification, and intervention at early disease stages are crucial to decrease mortality.
For decades, the complex communities of bacteria that colonize the gastrointestinal tract have been hypothesized to play an integral role in the pathophysiology of sepsis acting as the motor of sepsis and multiple organ dysfunction (4-6). However, only recently has this theory been supported by experimental evidence. The composition of the intestinal microbiome is severely affected by sepsis, which in turn might contribute to the development of organ failure (7,8). Knowledge of these specific microbiome changes during sepsis is therefore now recognized as a key question for intensive care research (9). Recent studies revealed that during sepsis, the gut is generally overrun by a single bacterial genus, including several pathogenic and antibiotic-resistant species such as Clostridium spp. and Enterococcus spp. (8). Moreover, the intestine of patients with sepsis loses key bacterial genera that represent an essential part of the microbiota of healthy individuals.
Therefore, we hypothesized a relationship between the gut microbiota and sepsis, and compared the gut microbiota in a cohort of children with sepsis and healthy volunteers through 16S rDNA gene sequencing. This study aimed to identify the gut microbiota profile of Chinese pediatrics sepsis and its correlation with their clinical features; as well as to assess whether the gut microbiota could serve as a potential prognostic marker for children with sepsis admitted into pediatric intensive care unit (PICU). We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/tp-21-51).
Methods
Subjects
A total of 25 children with sepsis were enrolled from PICU in Shanghai Children’s Medical Center from 2016 to 2018. Criteria for eligibility in the study included: (I) 28 days < the age ≤18 years, (II) critical illness requiring ICU admission, (III) diagnosis of sepsis according to the criteria of The Third Intestinal Consensus Definitions for Sepsis and Septic Shock (Sepsis-3) (1), and (IV) being able to obtain informed consent. The exclusion criteria included: (I) no fecal sample being withdrawn within 48 h after the diagnosis of sepsis, (II) patients in the neonatal period or those older than 18 years, (III) lack of informed consent. In addition to the sepsis group, 15 age- and sex-matched healthy children were selected as the control group. All healthy children did not have any disease that could potentially affect the stability of gut microbiota, such as diabetes, immunodeficiency, autoimmune diseases, inflammatory bowel disease or diarrhea in the past 3 months. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the Institutional Review Board and the Ethics Committee of Shanghai Children’s Medical Center (SCMCIRB-K2017066). Written informed consent was obtained from all participating patients and/or their parents.
Demographic and clinical data of the patients were recorded after ICU admission, including age, sex, vital signs, clinical presentations, application of antibiotics, and comorbidities. The assessment included routine laboratory tests such as lactic acid (LA), C-reactive protein (CRP), and procalcitonin (PCT), as well as the infection status determined within 24 h after the diagnosis of sepsis. Length of stay in ICU and clinical outcomes were recorded at the end of hospitalization.
Sample collection and DNA extraction
Fecal samples were collected from all the recruited subjects and stored at −80 °C until further processing. For 16S rDNA sequencing, genomic DNA was extracted from each fecal sample by the QIAamp Fast DNA Stool Mini Kit (Qiagen, Germany) according to the manufacturer’s protocol. The integrity and sizes of genomic DNA in each fecal sample were assessed using 1% agarose gel electrophoresis. The DNA was stored at −80 °C prior to sequencing.
16S rDNA amplification and sequencing
The composition of the gut microbiome in fecal samples was determined by 16S rDNA sequencing. Bacterial 16S rDNA gene libraires were generated independently for each sample by amplification with primers 515F (5'-GTGCCAGCMGCCGCGGTAA-3') and 806R (5'-GGACTAHVGGGTWTCTAAT-3'). The PCR conditions included an initial denaturation at 95 °C for 5 min, 20 cycles of denaturation at 98 °C for 20 s, annealing at 58 °C for 30 s, and elongation at 72 °C for 30 s, and a final extension at 72 °C for 5 min. PCR amplification was performed in triplicates using 50 µL reactions, with 10 pmol of forward and reverse primers and 100 ng of template. Amplicons were analyzed with 1.5% agarose gel electrophoresis, and the products with the desired size were purified using the QIAquick Gel Extraction Kit (QIAGEN, Germany). The products were submitted to the next-generation sequencing laboratory of Beijing Genomics Institute (BGI-Shenzhen, China), and 250 bp paired-end sequencing was performed on Illumina HiSeq 2500 platform.
Data analysis of 16S rDNA
Raw sequencing data were processed and analyzed using QIIME (Quantitative Insights Into Microbial Ecology, Version 1.9.1) software. Forward and reverse reads for each individual sample were demultiplexed, joined, and quality filtered. The reads were truncated at any site receiving an average quality score <20 over a 3 bp sliding window, and the truncated reads shorter than 75% of the raw reads were discarded. Sequences containing ambiguous characters or containing more than two nucleotide mismatches in the primer sequence were removed. Chimeric sequences were identified and removed from the dataset using the usearch61 method with reference data of the ‘gold’ set in http://sourceforge.net/projects/microbiomeutil/files/ (version 2011-11-02) (10). The “open-reference” QIIME protocol was used with the UCLUST method to select operational taxonomic units (OTUs) (11). Sequences with at least 97% similarity were clustered together, and representative sequence from each cluster was used to identify bacterial taxa from the Greengenes database as of August 2013. OTUs containing fewer than 2 sequences or with overall relative abundance <0.00005 were excluded from further analysis. QIIME was used to calculate alpha and beta diversity. Linear discriminant analysis (LDA) effect size (LEfSe) was used to identify different markers, and α=0.05 was used in Wilcoxon rank sum test, and the log value for LDA analysis was set to <2.0.
Statistical analysis
Continuous nonparametric data were analyzed using a Mann-Whitney U or Kruskal-Wallis test, and the results are presented as the median and interquartile range (IQR). Continuous parametric data presented as numbers and percentages or as the mean ± standard deviation (SD), were analyzed with a student’s t test or analysis of variance. Categorical data were analyzed using a Chi-square test. Receiver operating characteristics (ROC) curves were generated for variables including diversity, different phylum and their ratios; the Youden index (Youden index = sensitivity + specificity −1) were used to select the optimal cut-off value of the variables for the quantitative results of diagnostic tests as previously described (12). All data analyses were performed with R studio built under R version 3.2.3 and SPSS version 24.0. A P<0.05 was considered statistically significant. Figures were created using GraphPad Prism v.7.0 and Adobe Illustrator CC v.21.0.0.
Results
The primary characteristics of sepsis
The demographic characteristics of the sepsis and healthy controls were summarized in Table 1, and no differences in age or gender were detected between the two groups. All septic cases had different underlying diseases, including leukemia, aplastic anemia, congenital heart disease, biliary atresia, immunodeficiency, solid tumor, and Kawasaki disease. Of the 25 patients, 18 (72%) patients occurred septic shock, and 18 (72%) patients experienced single organ failure. Before fecal sample collection, all patients underwent antibiotic therapy, with the median therapeutic window of 9 days and average 4.4 different agents per person. Ten cases (40%) were positive for single pathogen as determined by blood culture, including 3 cases of Klebsiella pneumoniae, 3 cases of Acinetobacter baumannii, 2 cases of Staphylococcus aureus, 1 case of Streptococcus gallolyticus and 1 case of Pseudomonas aeruginosa. Mixed infections included 2 cases of Acinetobacter baumannii and Klebsiella pneumoniae, as well as 1 case of Escherichia coli and Klebsiella pneumoniae.
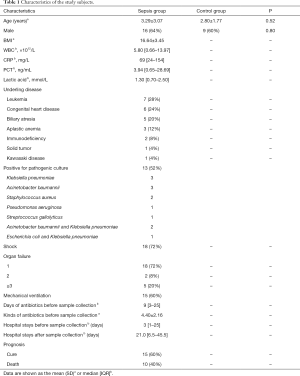
Full table
Microbiota analysis of the fecal sample in the sepsis and control groups
Alpha and beta diversity
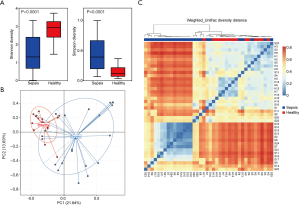
In the fecal microbiota, the mean community diversity index (i.e., α-diversity, including Shannon and Simpson based on OTU levels) of the sepsis group was significantly lower than that of the control group (Figure 1A), indicating that the richness of the gut microbiota in patients with sepsis was substantially lower compared with healthy children. A significant difference was also identified in β-diversity based on the weighted UniFrac between the two groups (Figure 1B and 1C), indicating that the fecal microbial community structure in the sepsis group was significantly different from the control group.
Alternation in the taxa
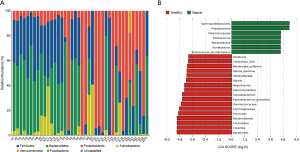
Effects on gut microbiota profile microbiota composition in children with sepsis was firstly evaluated at the phylum level. A relatively high abundance of Proteobacteria was observed in the sepsis group, while Firmicutes, Bacteroidetes and Actinobacteria were significantly higher in the control group (Figure 2A). Furthermore, a remarkable difference in fecal microbiota between the two groups was observed based on LEfSe analysis, indicating the different phylogenetic distribution of microbiota. At the genus level, the genera Roseburia, Bacteroides, Clostridia, Faecalibacterium, and Blautia were higher in healthy children. In contrast, the relative abundance of genera Acinetobacter and Enterococcus were higher in the patients with sepsis (Figure 2B).
Correlation analysis between changes of gut microbiota and clinical features in children with sepsis
Association between microbiota composition and sepsis infection status
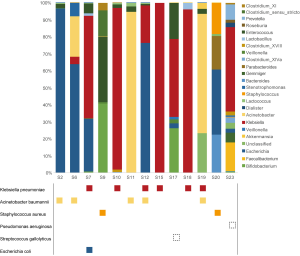
The abundant bacterial genus in the gut microbiota was found to correspond to the causative pathogen in sepsis (Figure 3). The pathogen identified by culture was one of the most abundances OTUs in the gut microbiota, with the corresponding genus being the most abundant gut bacterium in 6 cases (46.2% of culture-positive cases, including 4 cases of Klebsiella pneumoniae and 2 cases of Acinetobacter baumannii) and the second most abundant in 3 patients (23.1% of culture-positive cases, including 1 case of Escherichia coli, Klebsiella pneumoniae, and Acinetobacter baumannii, respectively).
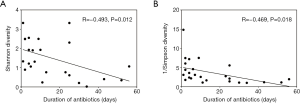
Association between alpha diversity and antibiotic usage
With the increasing duration of antibiotic usage before fecal sample collection, the diversity of gut microbiota decreased significantly (Figure 4), as evident by the negative correlation of Shannon index (R=–0.493, P=0.012) and the negative correlation of 1/Simpson index (R=–0.469, P=0.018) with the number of days of antibiotic usage, respectively. The duration of antibiotic usage was also positively correlated with the abundance of Acinetobacter baumannii (R=0.561, P=0.004) and Staphylococcus aureus (R=0.521, P=0.008) (data not shown). However, no correlation between the bacterial diversity with the antibiotic agent used was observed. Notably, children with sepsis who used the same antibiotic presented different gut microbiome patterns (Figure S1).
Changes of gut microbiota were associated with the prognosis of sepsis
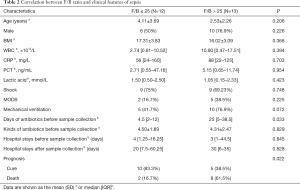
To explore whether microbiota-related markers could be related to the clinical feature of sepsis, ROC curve was generated and the Youden index was used to select the optimal cut-off value for the quantitative results of diagnostic tests (12). Among all the variables tested (including diversity, different phylum and their ratios), only the parameter of Firmicute/Bacteroidetes ratio (F/B-ratio), which is commonly used to describe the microbiota disorder, as well as the Bacteroidetes abundance, were correlated with sepsis prognosis. According to the ROC curve (Figure S2 and Table S1), the sepsis cohort was divided into two groups with low F/B-ratio (index ≤25) and high F/B-ratio (index >25). There were significantly intergroup differences in the antibiotic therapeutic window (P=0.033) and prognosis of sepsis (P=0.022; Table 2). Besides, a higher proportion in the mechanical ventilation (P=0.032) and death rate (P=0.048) were observed in patients with a low abundance of Bacteroidetes (index ≤5, Figure S3, Tables S2, S3).
Full table
Discussion
Several studies have shown that patients with sepsis have a profoundly distorted composition of the intestinal microbiota. Nevertheless, nearly all studies have focused on adults or neonates (7,13-15). Considering the heterogeneity of gut microbiota in different ages and races, the present study explored the relationship between gut microbiota and sepsis in Chinese children and analyzed the possibility of the gut microbiota as a prognostic biomarker of children with sepsis admitted into the PICU.
16S rDNA sequencing demonstrated that the richness of gut microbiota (indicated by α-diversity) in children with sepsis was significantly lower, and the community structure (shown by β-diversity index) in patients with sepsis was also significantly differed from that of healthy children. Such observation is consistent with a previous study by Lankelma et al., who reported a significantly lower bacterial diversity in patients with sepsis than the control subjects (P<0.0001) (13). Low-diversity microbiota has been observed in numerous pathogenic conditions, such as acute diarrheal disease (16), Clostridium difficile infection (CDI) (17), inflammatory bowel disease (IBD) (18), and obesity (19). The causal mechanisms underlying such low-diversity dysbiosis are probably the disruptive effects of the disease itself, as well as interventions during clinical care. Factors such as hypoxic injury and inflammation, intestinal dysmotility, disrupted epithelial integrity, shifts in intraluminal pH values, treatment with antibiotics, vasopressors, proton-pump inhibitors, opioids, and parenteral or enteral feeding, are all presented as potential disruptors of the microbiome in critical illness (20,21).
The use of broad-spectrum antibiotics potentially contributes to the significant disruptive effect among these interventions (22). However, the Surviving Sepsis Campaign (SSC) guidelines suggest that intravenous antibiotics should be given within one hour of severe sepsis. Thus, patients in ICU have often received the treatment of broad-spectrum antibiotics when highly suspected of sepsis. A prospective study on 1,265 ICUs across the world found that on any given day, 75% of patients received antibiotics (23). In our study, all children with sepsis were treated with antibiotics, varying from one to eight different types. With the increasing duration of antibiotic usage before sampling, the diversity of gut microbiota decreased significantly. A proof-of-principle intervention trial randomized 16 healthy young men to receive either broad-spectrum antibiotics (ciprofloxacin, vancomycin and metronidazole) or no treatment for 7 days. As expected, microbial diversity was significantly diminished after antibiotic treatment (24). However, no correlation between the bacterial diversity with the antibiotic agent was observed. In addition, patients who received the same level of antibiotics showed different gut microbiome patterns, and patients with similar gut microbiome patterns were treated with drastically different antibiotic regimens. This is consistent with Lankelma’s study, in which patients with sepsis were treated with a significantly higher number of antibiotic classes than those non-septic critically ill patients (P<0.01), but no differences in microbiota composition were observed between these two groups, at the phylum and genus levels (13). Therefore, antibiotic exposure cannot fully explain the changes in the gut microbiome of children with sepsis.
Among the pediatric sepsis in our study, it is worth noting the significant expansion of Proteobacteria compared to the healthy controls (P=0.0026). Proteobacteria are Gram-negative opportunistic pathogens that secrete potent inflammatory compounds such as lipopolysaccharide (LPS) (25). Numerous studies to date endorse the concept that a bloom of Proteobacteria in the gut reflects dysbiosis or unstable gut microbial community structure (26,27). The expanded abundance of Proteobacteria has been suggested as a diagnostic microbial signature of dysbiosis, epithelial dysfunction, and risk for disease development (28). Previous study demonstrated that during the course of sepsis, the gut was generally overrun by a single bacterial genus, including several pathogenic and antibiotic-resistant species (8), most of which were Proteobacteria. Consistently, our data also showed that Proteobacteria species, including Klebsiella pneumoniae, Acinetobacter baumannii and Escherichia coli dominated the gut microbiota in 9 patients with sepsis. Similarly, Lankelma et al. also showed a marked shift in fecal bacterial composition, with a single bacterial genus making up more than 50% of the gut microbiota (13). Interestingly, pathogens identified by culture corresponded to the most or second most abundant OTU in the gut. The underlying mechanism probably lies in that the gut epithelium becomes hyperpermeable during sepsis, which results in the translocation of gut flora into systemic circulation (29). Moreover, the intestinal of patients with sepsis in our study lost key bacterial genera that represent an important part of the microbiota of healthy individuals, including Faecalibacterium spp., Blautia spp., and Ruminococcaceae spp. These bacteria degrade complex plant polysaccharides that human cells cannot break down, and convert them into short-chain fatty acids (SCFAs), which have beneficial effects for both the gut integrity and systemic immunity in sepsis (30). Disappearance of Faecalibacterium spp., which has known anti-inflammatory properties, might further promote an unfavorable inflammatory state in the gut (31).
In addition to the signature of gut microbiota, its correlation with the clinical parameters of children with sepsis was analyzed. Our data showed that the increased F/B-ratio was associated with increased mortality, which might be used as a microbiota biomarker for prognosis prediction of sepsis. Early evidence suggested that the F/B-ratio was associated with survival in critically ill patients. However, the cut-off values were different for the correlation between gut microbiota and prognosis in the different disease models. Ojima et al. (7) reported that in ICU patients in the acute phase after admission, the B/F-ratio of >10 was seen in four of the six patients who died, whereas a B/F-ratio of <0.10 was seen in only one of the six deaths, and none of the survival had a B/F ratio of >10 or <0.10. It remains unclear how the change in F/B-ratio affects the prognosis of sepsis, which awaits further elucidation. Besides, future studies should explore whether the recovery of the extreme alterations in gut microbiota could improve sepsis prognosis.
This study presents some limitations. First, it is a preliminary study of children with sepsis in the Chinese Han nationality, and the sample size is relatively small. Second, we did not eliminate the substantial effect of antimicrobial therapy and other heterogeneity of patients with sepsis on the gut microbiota. Subsequent studies will attempt to increase sample size and design longitudinal follow-up to observe the dynamic changes of gut microbiota.
Conclusions
Overall, the present study provides evidence for gut microbiota disturbance in Chinese children with sepsis. The main finding, that changes in the gut microbiota were closely related to the clinical characteristics in sepsis, may improve our understanding of sepsis pathogenesis, and possibly improve microbiota-based diagnosis, monitoring and therapy for sepsis.
Acknowledgments
We would like to thank the patients and their parents for the support and cooperation in publishing this work. We would like to thank Dr. Haoyan Chen for assisting the data analysis.
Funding: This work was financially supported by National Natural Science Foundation of China (81971890 to XM, 81900812 to NS and 81701936 to YT), Shanghai Key Laboratory of Clinical Molecular Diagnostics for Pediatrics (20dz2260900), Shanghai Key Laboratory of Emergency Prevention, Diagnosis and Treatment of Respiratory Infectious Diseases (20dz2261100), Shanghai Municipal Health Commission (2019SY049 to XM), the Collaborative Innovation Center for Translational Medicine at Shanghai Jiao Tong University School of Medicine (TM201820 to QC), Clinical Science and Technology Innovation Project of Shanghai Hospital Development Center (SHDC12019X35 to QC), the Interdisciplinary Program of Shanghai Jiao Tong University (YG2016QN38 to YT and YG2017QN33 to NS), and Pudong New Area Science and Technology Development Fund (PKJ2018-Y43 to HR and PKJ2020-Y01 to YT).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/tp-21-51
Data Sharing Statement: available at http://dx.doi.org/10.21037/tp-21-51
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tp-21-51). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and was approved by the Research Ethics Committee of the Shanghai Children’s Medical Center of Shanghai Jiao Tong University (Ethical code: SCMCIRB-K2017066). Written informed consent was obtained from all participating patients and/or their parents.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016;315:801. [Crossref] [PubMed]
- Fleischmann C, Thomas–Rueddel DO, Hartmann M, et al. Hospital Incidence and Mortality Rates of Sepsis: An Analysis of Hospital Episode (DRG) Statistics in Germany From 2007 to 2013. Dtsch Aerzteblatt Online, 2016.
- Fleischmann-Struzek C, Goldfarb DM, Schlattmann P, et al. The global burden of paediatric and neonatal sepsis: a systematic review. Lancet Respir Med 2018;6:223-30. [Crossref] [PubMed]
- Klingensmith NJ, Coopersmith CM. The Gut as the Motor of Multiple Organ Dysfunction in Critical Illness. Crit Care Clin 2016;32:203-12. [Crossref] [PubMed]
- Clark JA, Coopersmith CM. INTESTINAL CROSSTALK. Shock 2007;28:384-93. [Crossref] [PubMed]
- Alverdy JC, Laughlin RS, Wu L. Influence of the critically ill state on host-pathogen interactions within the intestine: Gut-derived sepsis redefined. Crit Care Med 2003;31:598-607. [Crossref] [PubMed]
- Ojima M, Motooka D, Shimizu K, et al. Metagenomic Analysis Reveals Dynamic Changes of Whole Gut Microbiota in the Acute Phase of Intensive Care Unit Patients. Dig Dis Sci 2016;61:1628-34. [Crossref] [PubMed]
- Zaborin A, Smith D, Garfield K, et al. Membership and Behavior of Ultra-Low-Diversity Pathogen Communities Present in the Gut of Humans during Prolonged Critical Illness. mBio 2014;5:e01361-14 [Crossref] [PubMed]
- Schuijt TJ, van der Poll T, de Vos WM, et al. The intestinal microbiota and host immune interactions in the critically ill. Trends Microbiol 2013;21:221-9. [Crossref] [PubMed]
- Edgar RC, Haas BJ, Clemente JC, et al. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011;27:2194-200. [Crossref] [PubMed]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010;26:2460-1. [Crossref] [PubMed]
- Hajian-Tilaki K. The choice of methods in determining the optimal cut-off value for quantitative diagnostic test evaluation. Stat Methods Med Res 2018;27:2374-83. [Crossref] [PubMed]
- Lankelma JM, van Vught LA, Belzer C, et al. Critically ill patients demonstrate large interpersonal variation in intestinal microbiota dysregulation: a pilot study. Intensive Care Med 2017;43:59-68. [Crossref] [PubMed]
- Stewart CJ, Embleton ND, Marrs ECL, et al. Longitudinal development of the gut microbiome and metabolome in preterm neonates with late onset sepsis and healthy controls. Microbiome 2017;5:75. [Crossref] [PubMed]
- Berkhout DJC, Niemarkt HJ, de Boer NKH, et al. The potential of gut microbiota and fecal volatile organic compounds analysis as early diagnostic biomarker for necrotizing enterocolitis and sepsis in preterm infants. Expert Rev Gastroenterol Hepatol 2018;12:457-70. [Crossref] [PubMed]
- David LA, Weil A, Ryan ET, et al. Gut Microbial Succession Follows Acute Secretory Diarrhea in Humans. Relman DA, editor. MBio, 2015;6.
- van Nood E, Vrieze A, Nieuwdorp M, et al. Duodenal Infusion of Donor Feces for Recurrent Clostridium difficile. N Engl J Med 2013;368:407-15. [Crossref] [PubMed]
- Lozupone CA, Stombaugh J, Gonzalez A, et al. Meta-analyses of studies of the human microbiota. Genome Res 2013;23:1704-14. [Crossref] [PubMed]
- Gérard P. Gut microbiota and obesity. Cell Mol Life Sci 2016;73:147-62. [Crossref] [PubMed]
- Haak BW, Wiersinga WJ. The role of the gut microbiota in sepsis. Lancet Gastroenterol Hepatol 2017;2:135-43. [Crossref] [PubMed]
- Dickson RP. The microbiome and critical illness. Lancet Respir Med 2016;4:59-72. [Crossref] [PubMed]
- Blaser M. Stop the killing of beneficial bacteria. Nature 2011;476:393-4. [Crossref] [PubMed]
- Vincent JL. International Study of the Prevalence and Outcomes of Infection in Intensive Care Units. JAMA 2009;302:2323. [Crossref] [PubMed]
- Lankelma JM, Cranendonk DR, Belzer C, et al. Antibiotic-induced gut microbiota disruption during human endotoxemia: a randomised controlled study. Gut 2017;66:1623-30. [Crossref] [PubMed]
- Shin NR, Whon TW, Bae J-W. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol 2015;33:496-503. [Crossref] [PubMed]
- Karlsson FH, Tremaroli V, Nookaew I, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 2013;498:99-103. [Crossref] [PubMed]
- Faith JJ, Guruge JL, Charbonneau M, et al. The Long-Term Stability of the Human Gut Microbiota. Science 2013;341:1237439 [Crossref] [PubMed]
- Litvak Y, Byndloss MX, Tsolis RM, et al. Dysbiotic Proteobacteria expansion: a microbial signature of epithelial dysfunction. Curr Opin Microbiol 2017;39:1-6. [Crossref] [PubMed]
- Jones WG, Minei JP, Barber AE, et al. Bacterial Translocation and Intestinal Atrophy After Thermal Injury and Burn Wound Sepsis. Ann Surg 1990;211:399-405. [Crossref] [PubMed]
- Rajilić-Stojanović M, de Vos WM. The first 1000 cultured species of the human gastrointestinal microbiota. FEMS Microbiol Rev 2014;38:996-1047. [Crossref] [PubMed]
- de Vos WM, de Vos EA. Role of the intestinal microbiome in health and disease: from correlation to causation. Nutr Rev 2012;70:S45-S56. [Crossref] [PubMed]


