
Characteristics of the intestinal microbiota and metabolism in infants with extrauterine growth restriction
Introduction
Extrauterine growth restriction (EUGR) is a clinical problem in which the growth values in affected neonates are lower than the tenth percentile of intrauterine growth expectation in preterm neonates at the time of discharge (1-3). Previous studies have put forth the idea that EUGR may induce adverse health outcomes such as impaired neurodevelopment (4) and metabolic syndromes (5). Although the factors leading to EUGR are complicated, one of the most principal may be energy deficiency (6). Regardless of the growing amount of ameliorative treatment and care regimens, it is important to understand the mechanisms underlying EUGR.
Multiple studies have reported on the interactions of gut microbiota alterations and adverse health states. The intestinal microbiota consists of trillions of microorganisms inhabiting the intestinal tract of the hosts and working as a metabolic organ. Though the mechanism is still under intensive study, the intestinal microbiota could influence the physical status of the hosts though several small molecules, such as the metabolites it generates in the gut (7). Thus, the intestinal microbiota plays essential roles in appetite, gut motility, energy uptake and storage, immunity, and protection against pathogens of the hosts (8-11). Proper microbiome or metabolome biomarkers may help in understanding the mechanism and diagnosing infants with EUGR.
By utilization of the metagenome together with nontargeted liquid chromatography-mass spectrometry (LC-MS) metabolomics, our knowledge of the gut microbiota has been greatly extended. In the present study, 50 EUGR infants and the 23 non-EUGR infants were enrolled. The gut microbiota, as well as the metabolites in feces and blood were examined to identify potential biomarkers. Any correlations between differential species and metabolites were evaluated to understand further the relationship between alterations in the microbiota and the metabolism status of the host.
We present the following article in accordance with the MDAR reporting checklist (available at: http://dx.doi.org/10.21037/tp-20-431).
Methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Research Ethics Committee of Shanghai Jiao Tong University Affiliated Sixth People’s Hospital (2017-009) and informed consent was taken from all the patients.
Study cohort
This retrospective study was carried out by reviewing the clinical charts of 73 preterm infants in the Shanghai Jiao Tong University Affiliated Sixth People’s Hospital. The inclusion criteria were: (I) infants with all neonatal curves below the 10th percentile of infants in the same gestational age (GA) when they were discharged from hospital; (II) written informed consent was obtained from the parents. The exclusion criteria were: (I) born with severe abnormalities, congenital heart or pulmonary disease, severe digestive tract malformation, metabolic disease or chromosomal disorders; (II) dead during hospitalization or with poor comorbid disease control; (III) mandatory discharge from the hospital without meeting the standard required.
Fecal collection and microbiome analysis
The fecal samples were collected in sterile containers and kept at −20 °C until further examination. The genomic DNA was purified with a QIAamp PowerFecal DNA Kit following the instructions of the manufacturer.
Extraction of metabolites from feces and blood
The fecal samples and blood samples were collected in sterile containers and kept at −20 °C until further examination.
For the fecal samples, 100 mg was transferred into 2-mL centrifuge tubes along with 500 µL 4 °C ddH2O. The centrifuge tubes were vortexed for 60 s. Then, 1,000 µL methanol (pre-cooled at −20 °C) was added and vortexed for 30 s. The tubes were placed into an ultrasound machine at room temperature for 10 min and then stewed for 30 min on ice. The tubes were centrifuged for 10 min at 14,000 rpm and 4 °C. Next, 1.2 mL supernatant was transferred into a new centrifuge tube and blow-dried by vacuum concentration.
For the blood samples, 100 µL of each sample was transferred into a 1.5-mL centrifuge tube along with 400 µL methanol (pre-cooled at −20 °C) and vortexed for 60 s. Then, the samples were centrifuged at 4°C and 12,000 rpm for 10 min. The supernatant of each sample was transferred into a new 1.5-mL tube. After centrifugation, the samples were blow-dried by vacuum concentration.
All the dried samples were dissolved with 150 µL of a 2-chlorobenzalanine (4 ppm) methanol aqueous solution (4:1, 4 °C) and filtered with a 0.22-µm membrane. Then, the samples were ready for LC-MS.
Metabolome analysis with LC-MS
Chromatographic separation was accomplished in a Thermo Ultimate 3000 system equipped with an ACQUITY UPLC® HSS T3 Column (150×2.1 mm, 1.8 µm, Waters) maintained at 40 °C. The temperature of the autosampler was 8 °C. Gradient elution of the analytes was carried out with 0.1% formic acid in acetonitrile (C) and 0.1% formic acid in water (D) or acetonitrile (A) and 5 mM ammonium formate in water (B) at a flow rate of 0.25 mL/min. The injection of 2 µL of each sample was done after equilibration.
The ESI-MSn experiments were executed on a Thermo Q Exactive Focus mass spectrometer with a spray voltage of 3.8 kV and −2.5 kV in the positive and negative modes, respectively. The sheath gas and auxiliary gas were set at 45 and 15 arbitrary units, respectively. The capillary temperature was 325 °C. The Orbitrap analyzer scanned over a mass range of m/z 81–1,000 for a full scan at a mass resolution of 70,000. Data-dependent acquisition MS/MS experiments were performed with HCD scanning. The normalized collision energy was 30 eV. Dynamic exclusion was implemented to remove some unnecessary information from the MS/MS spectra.
Statistical analyses
SPSS 23 was used to analyze the data, which are presented as the mean ± SE. The Wilcoxon rank sum test was employed for comparisons between groups with a non-normal distribution. Correlations between variables were evaluated using the Spearman coefficient. A P value <0.05 was considered to be a statistically significant finding.
Results
Study population
Table 1 shows the baseline of the study cohorts. In detail, we enrolled 50 infants with EUGR and 23 infants without EUGR between December 2017 and July 2018.
Full table
Overview of the different intestinal bacterial species between the EUGR and non-EUGR groups
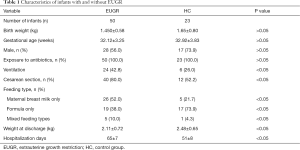
The microbial composition was detected by metagenome sequencing (12). As shown in Figure 1, the intestinal microbiota of the EUGR group exhibited a lower level of Bacteroides vulgatus (P<0.05), Dorea unclassified (P<0.05), Lachnospiraceae bacterium 1_1_57FAA (P<0.001), and Roseburia unclassified (P<0.05) than that of the non-EUGR group. In fact, L. bacterium 1_1_57FAA was absent in the EUGR group. Meanwhile, the EUGR group showed a higher level of Streptococcus mitis_oralis_pneumoniae (P<0.05). These results indicate that L. bacterium 1_1_57FAA could distinguish infants with or without EUGR.
Overview of the different Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways between the EUGR and non-EUGR groups
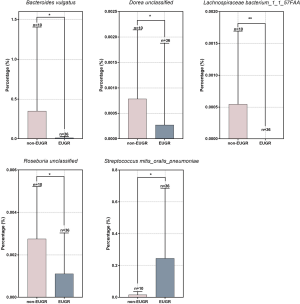
To investigate the potentially influenced metabolic pathways induced by EUGR status, we further analyzed the metagenome sequencing readings with KEGG. As shown in Figure 2, there were seven different enriched KEGG pathways between the non-EUGR group and EUGR group. Among these pathways, the non-EUGR group exhibited significantly higher levels of bile secretion (P<0.001), galactose metabolism (P<0.05), glycine, serine, and threonine metabolism (P<0.05), and the renin-angiotensin system (P<0.05) compared with the EUGR group. However, the EUGR group exhibited significantly higher levels of the cAMP signaling pathway (P<0.05), yeast meiosis (P<0.05), and oxidative phosphorylation (P<0.05) in comparison to the non-EUGR group.
Overview of the different metabolites between the EUGR and non-EUGR groups
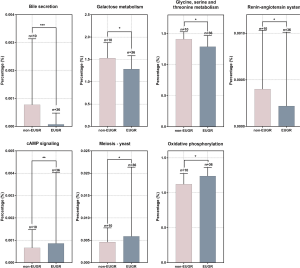
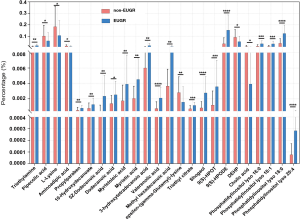
The metabolites generated by the intestinal microbiota affect the biological processes of the hosts, including appetite control and weight management. As infants with EUGR exhibit a lower growth velocity, we sought to examine the metabolites in the feces and blood of the infants of both groups. A non-targeted analysis of the metabolite composition was carried out to analyze the different metabolites between the infants with EUGR and non-EUGR. Figures 3 and 4 show the different metabolites in the blood and feces, respectively.
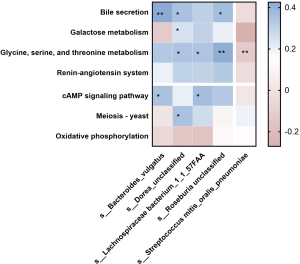
Correlation between the different microbial species and different KEGG pathways
To investigate the association between the different species and KEGG pathways, Spearman’s correlation analysis was used. As presented in Figure 5, among the KEGG pathways higher in the non-EUGR group, bile secretion was positively correlated with B. vulgatus (r=0.43, P<0.01), D. unclassified (r=0.31, P<0.05), and R. unclassified (r=0.33, P<0.05). Galactose metabolism was positively correlated with D. unclassified (r=0.14, P<0.05). Glycine, serine, and threonine metabolism was positively correlated with D. unclassified (r=0.30, P<0.05), L. bacterium 1_1_57FAA (r=0.30, P<0.05), and R. unclassified (r=0.40, P<0.01). Among the KEGG pathways higher in the EUGR group, the cAMP signaling pathway was positively correlated with B. vulgatus (r=0.31, P<0.05) and L. bacterium 1_1_57FAA (r=0.33, P<0.05). Yeast meiosis was positively correlated with D. unclassified (r=0.31, P<0.05). In brief, the different intestinal microbial species were significantly correlated with the metabolic pathways. These results indicate that these intestinal microbial species may play a role in the metabolism of the hosts.
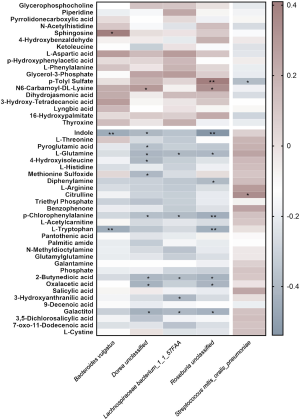
Correlation between the blood metabolome and intestinal microbiota
To evaluate the association between the different blood metabolites and different Microbial species, Spearman’s correlation was analyzed for the five different species and 48 different serum metabolites; the results are graphically presented in Figure 6. In general, a total of 30 significant interactions were found. Among the total different serum metabolites, 16 of them were significantly correlated with different Microbial species, half of which were amino acids and the rest other metabolites. Among the microbial species enriched in the non-EUGR group, B. vulgatus was negatively correlated with indole (r=−0.43, P<0.01) and L-tryptophan (r=−0.43, P<0.01), which were much higher in the EUGR group. Meanwhile, it was positively correlated with sphingosine (r=0.41, P<0.05), which was higher in the non-EUGR group. Additionally, D. unclassified was negatively correlated with indole (r=−0.37, P<0.05), L-glutamine (r=−0.38, P<0.05), pyroglutamic acid (r=−0.32, P<0.05), 4-hydroxyisoleucine (r=−0.40, P<0.05), methionine sulfoxide (r=−0.37, P<0.05), p-chlorophenylalanine (r=−0.33, P<0.05), 2-butynedioic acid (r=−0.37, P<0.05), oxaloacetic acid (r=−0.39, P<0.05), and galactitol (r=−0.40, P<0.05), which were all more abundant in the EUGR group. Moreover, it was positively correlated with N6-carbamoyl-DL-lysine (r=0.32, P<0.05), which was more abundant in the non-EUGR group. Furthermore, L. bacterium 1_1_57FAA was negatively correlated with L-glutamine (r=−0.36, P<0.05), p-chlorophenylalanine (r=−0.35, P<0.05), 2-butynedioic acid (r=−0.35, P<0.05), 3-hydroxyanthranilic acid (r=−0.37, P<0.05), and galactitol (r=−0.37, P<0.05), which were more abundant in the EUGR group. In addition, R. unclassified was negatively correlated with indole (r=−0.54, P<0.01), L-glutamine (r=−0.37, P<0.05), diphenylamine (r=−0.36, P<0.05), p-chlorophenylalanine (r=−0.42, P<0.01), L-tryptophan (r=−0.41, P<0.01), 2-butynedioic acid (r=−0.38, P<0.05), oxaloacetic acid (r=−0.38, P<0.05), and galactitol (r=−0.40, P<0.05), which were more abundant in the EUGR group. Meanwhile, it was positively correlated with p-tolyl sulfate (r=0.41, P<0.01) and N6-carbamoyl-DL-lysine (r=0.36, P<0.05), which were more abundant in the non-EUGR group. Finally, S. mitis_oralis_pneumoniae, which exhibited much higher levels in the EUGR group, was positively correlated with citrulline (r=0.36, P<0.05) and negatively correlated with p-tolyl sulfate (r=−0.35, P<0.05).
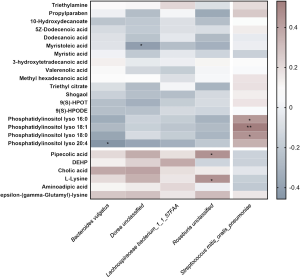
Correlation between the fecal metabolome and gut microbiota
To address the correlations between the different fecal metabolites and the different gut microbiota, Spearman’s correlation was calculated for the five different microbial species and 24 different fecal metabolites. As shown in Figure 7, a total of seven interactions with statistical significance were found. For instance, R. unclassified was positively correlated with pipecolic acid (r=0.45, P<0.05) and L-lysine (r=0.45, P<0.05), both of which were much higher in the non-EUGR group. Moreover, B. vulgatus was negatively correlated with phosphatidylinositol lyso 20:4 (r=−0.46, P<0.05), and D. unclassified was negatively correlated with myristoleic acid (r=−0.42, P<0.05). However, S. mitis_oralis_pneumoniae, which exhibited much higher levels in the EUGR group, was positively correlated with phosphatidylinositol lyso 16:0 (r=0.42, P<0.05), phosphatidylinositol lyso 18:1 (r=0.53, P<0.05), and phosphatidylinositol lyso 18:0 (r=0.44, P<0.05), which were higher in the fecal from the EUGR group.
Discussion
This study aimed to identify potential biomarkers for distinguishing infants with EUGR from infants without EUGR through gut microbiotas and metabolites. Here, we observed four lower-gut microbial species, B. vulgatus (13-15), D. unclassified, R. unclassified, and L. bacterium 1_1_57FAA, in the intestinal microbiotas of infants with EUGR compared with those of infants without EUGR. Although the functions of these species have not been well characterized, our results suggest that these species are relevant to the EUGR problem. Meanwhile, we observed that S. mitis_oralis_pneumoniae (16), which are regarded as common human pathogens, were higher in the gut microbiota of the EUGR group than in that of the non-EUGR group. Taken together, our results suggest a specific intestinal microenvironment of infants with EUGR.
As a metabolic organ, it is possible for different intestinal microbiotas to contribute to different metabolites in hosts, and vice versa. Our results exhibit that the different microbial species were correlated with KEGG metabolic pathways. Thus, it was reasonable to find that the different intestinal microbial species were associated with different metabolites. Our study demonstrated significantly higher levels of several individual amino acids and organic acids in the blood of the EUGR group than in that of the non-EUGR group. Specifically, the higher blood level of L-tryptophan was positively correlated with B. vulgatus and R. unclassified. Meanwhile, the higher blood level of indole was positively correlated with B. vulgatus, D. unclassified, and R. unclassified. Tryptophan is an essential amino acid obtained from the daily diet, and it participates in several metabolic pathways. It has been reported to modulate the gut microbiota and, thus, protect the gut barrier. However, only 4–6% of the total tryptophan can be metabolized by the gut microbiota and generate indole, which may be toxic to the hosts (17). We observed a high level of indole in blood of the EUGR group, which indicates a dysfunction of the gut barrier and a leakage of harmful substances into the blood. In conclusion, B. vulgatus, D. unclassified, and R. unclassified may play a possible protective role in gut homeostasis, which is probably involved in the energy deficiency of infants with EUGR.
Furthermore, our results showed four individual species of lysophosphatidylinositol (LPI), which were significantly more abundant in the fecal samples from the EUGR group as compared with those from the non-EUGR group. Notably, phosphatidylinositol lyso 16:0, 18:1, and 18:0 were positively correlated with S. mitis_oralis_pneumoniae. Meanwhile, phosphatidylinositol lyso 20:4 was negatively correlated with B. vulgatus. An LPI is generated by the enzymatic activity of PLA2, which hydrolyzes glycerophospholipids and sphingolipids. Both plasma LPIs and serum LPIs have been reported to be associated with obesity, type 2 diabetes, and several types of cancer (18). So far, however, the roles of fecal LPIs have rarely been reported (19). Fecal LPIs could be probably derived from the undigested LPIs in food, which indicates lower efficiency of the digestive system in EUGR groups. Alternatively, fecal LPIs could be the intermediate metabolites in phospholipid homeostasis. Despite our inability to point out the roles of fecal LPIs in a physiological context, our results exhibit a correlation between fecal LPIs and different microbial species. In short, fecal LPIs could be represented as useful biomarkers for infants with EUGR.
In summary, as gut microbiota homeostasis exerts multiple beneficial effects on the microbiota-derived metabolites and energy uptake of humans, the dysregulation of infant gut microbiota may promote EUGR. Thus, it is crucial to identify the biomarkers of EUGR in infants. Ongoing research is needed to gain a better understanding of the mechanisms involved in the regulation of metabolites by EUGR-specific microbial species.
Acknowledgments
Funding: The work was supported by the people’s livelihood research special funding of Shanghai Science and Technology Development (grant number: PKJ2017-Y09).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tp-20-431
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tp-20-431
Peer Review File: Available at http://dx.doi.org/10.21037/tp-20-431
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tp-20-431). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Research Ethics Committee of Shanghai Jiao Tong University Affiliated Sixth People’s Hospital (2017-009) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Clark RH, Thomas P, Peabody J. Extrauterine growth restriction remains a serious problem in prematurely born neonates. Pediatrics 2003;111:986-90. [Crossref] [PubMed]
- Astbury J, Orgill AA, Bajuk B, et al. Sequelae of growth failure in appropriate for gestational age, very low-birthweight infants. Dev Med Child Neurol 1986;28:472-9. [Crossref] [PubMed]
- Ruth VA. Extrauterine growth restriction: a review of the literature. Neonatal Netw 2008;27:177-84. [Crossref] [PubMed]
- Chien HC, Chen CH, Wang TM, et al. Neurodevelopmental outcomes of infants with very low birth weights are associated with the severity of their extra-uterine growth retardation. Pediatr Neonatol 2018;59:168-75. [Crossref] [PubMed]
- Parkinson JR, Hyde MJ, Gale C, et al. Preterm birth and the metabolic syndrome in adult life: a systematic review and meta-analysis. Pediatrics 2013;131:e1240-63. [Crossref] [PubMed]
- Su BH. Optimizing nutrition in preterm infants. Pediatr Neonatol 2014;55:5-13. [Crossref] [PubMed]
- Zierer J, Jackson MA, Kastenmüller G, et al. The fecal metabolome as a functional readout of the gut microbiome. Nat Genet 2018;50:790-5. [Crossref] [PubMed]
- Leser TD, Mølbak L. Better living through microbial action: the benefits of the mammalian gastrointestinal microbiota on the host. Environ Microbiol 2009;11:2194-206. [Crossref] [PubMed]
- Vasaï F, Ricaud KB, Cauquil L, et al. Lactobacillus sakei modulates mule duck microbiota in ileum and ceca during overfeeding. Poult Sci 2014;93:916-25. [Crossref] [PubMed]
- Whiteside SA, Razvi H, Dave S, et al. The microbiome of the urinary tract--a role beyond infection. Nat Rev Urol 2015;12:81-90. [Crossref] [PubMed]
- Heiss CN, Olofsson LE. Gut Microbiota-Dependent Modulation of Energy Metabolism. J Innate Immun 2018;10:163-71. [Crossref] [PubMed]
- Wang Y, Zhou Y, Xiao X, et al. Metaproteomics: A strategy to study the taxonomy and functionality of the gut microbiota. J Proteomics 2020;219:103737 [Crossref] [PubMed]
- Ó Cuív P, de Wouters T, Giri R, et al. The gut bacterium and pathobiont Bacteroides vulgatus activates NF-κB in a human gut epithelial cell line in a strain and growth phase dependent manner. Anaerobe 2017;47:209-17. [Crossref] [PubMed]
- Nicolucci AC, Hume MP, Martínez I, et al. Prebiotics Reduce Body Fat and Alter Intestinal Microbiota in Children Who Are Overweight or With Obesity. Gastroenterology 2017;153:711-22. [Crossref] [PubMed]
- Qi X, Yun C, Sun L, et al. Gut microbiota-bile acid-interleukin-22 axis orchestrates polycystic ovary syndrome. Nat Med 2019;25:1225-33. Erratum in: Nat Med 2019;25:1459. [Crossref] [PubMed]
- Chen JH, She KK, Wong OY, et al. Use of MALDI Biotyper plus ClinProTools mass spectra analysis for correct identification of Streptococcus pneumoniae and Streptococcus mitis/oralis. J Clin Pathol 2015;68:652-6. [Crossref] [PubMed]
- Ridlon JM, Kang DJ, Hylemon PB, et al. Bile acids and the gut microbiome. Curr Opin Gastroenterol 2014;30:332-8. [Crossref] [PubMed]
- Piñeiro R, Falasca M. Lysophosphatidylinositol signalling: new wine from an old bottle. Biochim Biophys Acta 2012;1821:694-705. [Crossref] [PubMed]
- Inoue M, Adachi M, Shimizu Y, et al. Comparison of lysophospholipid levels in rat feces with those in a standard chow. J Agric Food Chem 2011;59:7062-7. [Crossref] [PubMed]


