
Highly homogeneous microbial communities dominated by Mycoplasma pneumoniae instead of increased resistance to macrolide antibiotics is the characteristic of lower respiratory tract microbiome of children with refractory Mycoplasma pneumoniae pneumonia
Introduction
Mycoplasma pneumoniae (M. pneumoniae) is an important pathogen of community-acquired pneumonia in children. For the past few years, because of the emergence of M. pneumoniae resistance to macrolides and a series of complex mechanisms, the incidence of refractory M. pneumoniae pneumonia (RMPP) has increased, often leading to poor prognosis and sequelae such as atelectasis, pulmonary necrosis, bronchiectasis, and bronchiolitis obliterans (1-3).
Researchers found that the microbiota changed significantly during the lower respiratory tract (LRT) infection and was closely related to the prognosis or course of pneumonia (4,5). However, the structure and function of LRT microbiota in patients with RMPP remained unexplored. We reasoned that characterization of LRT microbiota in children with RMPP may provide us with opportunities to uncover the ecosystem of the LRT, which can lead to the mechanism of aggravation of non-refractory M. pneumoniae pneumonia (NRMPP) to RMPP, and possible avenues for the prevention of RMPP.
Here, a prospective study was conducted in the Children’s Hospital of Fudan University to characterize the microbiota in LRT of RMPP and NRMPP children as well as its function and to reveal the correlation between microbiota and laboratory tests.
We present the following article in accordance with the MDAR checklist (available at http://dx.doi.org/10.21037/tp-20-404).
Methods
Patient enrollment and study design
During the period between June 28, 2019 and March 23, 2020, we prospectively enrolled 52 children admitted to the hospitalization zone in the Division of Pulmonary Medicine, Children’s Hospital of Fudan University, with clinically diagnosed NRMPP or RMPP, and collected bronchoalveolar lavage fluid (BALF) when bronchoscopy was necessary. The inclusion criteria were (I) diagnosed with pneumonia and meet the hospitalization standard of community-acquired pneumonia in children (has one of the following): Under the condition of breathing air, RR >50 times/min; under the condition of breathing air, SaO2 ≤0.92; dyspnea: chest wall inspiratory depression, flaring of alaenasi; intermittent apnea; persistent high fever for 3-5 days; chest X-ray and other imaging data confirmed bilateral or multiple lobes involvement or atelectasis, pleural effusion or short-term progression (6); (II) M. pneumoniae infection, which was confirmed by conducting a serological test (M. pneumoniae-IgM-positive and an antibody titer ≥1:160 or a ≥4-fold increase in the titer) and by detection of M. pneumoniae nucleic acid in the nasopharyngeal aspirate, sputum and BALF (7); (III) bronchoscopy, which was routinely performed when patients had more than one lobe of pulmonary consolidation or atelectasis by X-ray or CT scan. RMPP was defined as clinical symptoms and the pulmonary imaging of the pneumonia aggravated after regular treatment with macrolide antibiotics for more than seven days. The exclusion criteria were (I) the detection of any other pathogens in the patients’ blood, nasopharyngeal aspirate, sputum or BALF via culture, viral antigen detection assays (adenovirus, respiratory syncytial virus, metapneumovirus, influenza, and parainfluenza), interferon-γ release assays, and T cell spot tests (T-SPOTs); (II) children who combined with other respiratory diseases, blood system disease, autoimmune disease or with organ dysfunction and in the acute stage of allergic disease were also excluded.
The ethical application and the consent procedure of this study were approved by the Ethics Committee of Children’s Hospital of Fudan University (No. 2016-87). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013), and informed consent was taken from all the parents.
Clinical characteristics
Clinical information was collected from the medical records of the patients, and the age, sex, hospitalization days and antibiotic treatments before bronchoscopy were recorded. The maximum levels of the laboratory data were recorded before bronchoscopy was performed, which included white blood cell (WBC) counts, levels of C-reactive protein (CRP), fibrinogen, lactate dehydrogenase (LDH) and D-dimer.
Sample collection
Bronchoscopy was done by using standard methods and conscious intravenous sedation with midazolam. After topical anesthesia of the larynx, trachea, and carina was achieved with 2% lidocaine (Sanchine, China), the bronchoscope (Olympus, Japan) was wedged in the lesion’s segment or lobe and BALF was performed with three aliquots of sterile saline (Baxter, China), 1 ml/kg each, followed by suction pressure of 100 mmHg. All BALF samples were then immediately frozen in a dry liquid nitrogen Dewar (ASONE, Japan) and stored at −80 °C until analysis.
Nucleic acid extraction
BALF samples were collected as described above. DNA was extracted from BALF, using the TIANamp Magnetic DNA Kit (Tiangen, China) according to the manufacturer’s protocols. The quantity and quality of DNA was assessed using the Qubit (Thermo Fisher Scientific, USA) and the NanoDrop (Thermo Fisher Scientific, USA), respectively. No template control (NTC) of each batch were also processed with clinical samples.
Library preparation and sequencing
DNA libraries were prepared using the KAPA Hyper Prep kit (KAPA Biosystems, USA) according to the manufacturer’s protocols, by fragmenting DNA into 150–300 bp. DNA libraries were qualified by Agilent 2100, and 75 bp single-ends sequenced on Illumina NextSeq 550Dx (Illumina, USA).
Bioinformatics analysis
Raw sequencing data was split by bcl2fastq2, and high-quality sequencing data were generated using Trimmomatic by removing low quality reads, adapter contamination, and duplicated and short (length <35 bp) reads. Human host sequences were subtracted by mapping to human reference genome (hs37d5) using bowtie2. Reads that could not be mapped to the human genome were retained and aligned with microorganism genome database for microbial identification by Kraken, and for species abundance estimation by Bracken. The microorganism genome database contained genomes or scaffolds of bacteria, fungi, viruses, and parasites (download from ftp://ftp.ncbi.nlm.nih.gov/genomes/genbank/). We used the following criteria for positive results of metagenomic next generation sequencing (mNGS): For Mycobacterium, Nocardia and Legionella pneumophila, the result was considered positive if a species detected by mNGS had a species-specific read number ≥1. For bacteria (excluding Mycobacterium, Nocardia and Legionella pneumophila), fungi, virus and parasites, the result was considered positive if a species detected by mNGS had at least 3 non-overlapping reads. Pathogens detected in the negative NTC were excluded but only if the detected reads were ≥10-fold than that in the NTC. Functional metagenome profiling was calculated using HUMAnN2 standard workflow with default settings. Tables of normalized gene family were converted into KEGG Orthology terms (KO) using humann2_regroup_table command of HUMAnN2. Normalized KO were then mapped to KEGG pathways and KEGG modules using MinPath, as provided with the HUMAnN2. Finally, the profiles of species and KEGG pathways were retained for further analysis if they showed non-zero abundance in at least 40% of samples.
DIAMOND was used in mapping CARD and Virulence Factor Database (VFDB) to identify resistance genes and virulence factors.
Draft genome of M. pneumoniae
Host-removed reads from all samples were co-assembled using megahit. Contigs of all samples were clustered to recover metagenome-assembled genomes (MAGs) using MetaBAT2 under default parameters. The quality of MAGs was assessed using CheckM and QUAST. The taxonomic classification of MAGs was performed based on whole-genome alignment against the genome sequences published by IMG, NCBI, and PATRIC. Finally, we obtained the draft genome of M. pneumoniae, and gene prediction of M. pneumoniae was performed using MetaGeneMark, followed by identification of non-redundant genes using CD-HIT. The annotations of antibiotic resistance and virulence genes were acquired through CARD and VFDB using BLASTP.
Statistical analysis
The statistical analyses of clinical records were performed using SPSS software (IBM, version 20.0). Data with a skewed distribution were presented as median values (interquartile ranges: Q1, Q3). The different groups were compared using the Mann-Whitney U test. The statistical significance of the differences in the categorical variables was determined using the chi-square test. Statistical significance was determined at the two-tailed 0.05 level. Other statistical analyses were performed by R software (v4.0.1). Alpha diversity was estimated by the Shannon and Simpson index based on the taxonomic profile of each sample, beta diversity was assessed by the Bray-Curtis measure, and compared between NRMPP and RMPP patients by using Wilcoxon rank sum test, and was subsequently visualized by principal coordinate analysis (PCoA) plot. Permutational multivariate analysis of variance (PERMANOVA) was performed by the R package “vegan” to analyze Bray-Curtis distance in different NRMPP and RMPP groups. Differential relative abundance of taxonomic groups at the genus level was tested by using Kruskal-Wallis rank sum test (R package “kruskal.test”). The genera with mean relative abundances greater than 0.4% and penetrance greater than 40% among all samples were compared. Spearman’s correlations between clinical characteristics and the relative abundances of genera were calculated by the R package “cor.test”, and FDR correction was adopted to adjust all P values. Statistically significant differences in the relative abundance of microbe and KEGG pathways among groups were assessed by the linear discriminant analysis of effect size (LEfSe) analysis.
Results
Demographical characteristics
In this study, the NRMPP group comprised 9 males and 18 females, and the RMPP group comprised 11 males and 14 females. The characteristics of these subjects are summarized in Table 1. No significant differences were observed between the baseline characteristics of the two groups (P>0.05). There was no difference in the treatment of methylprednisolone among groups (P>0.05). As for antibiotics treatment, no significant (P>0.05) difference in treatment of azithromycin, levofloxacin, ciprofloxacin, and minocycline was observed whereas a significant difference was observed in the treatment of ceftriaxone, cefoperazone/sulbactam and clarithromycin (P<0.05). Regarding laboratory data and hospitalizations days, higher LDH, D-dimer levels and prolonged hospitalization days were observed in the RMPP group than NRMPP group (P<0.05), whereas markers of systemic inflammation such as WBC counts, CRP and fibrinogen levels had no difference between groups (P>0.05).
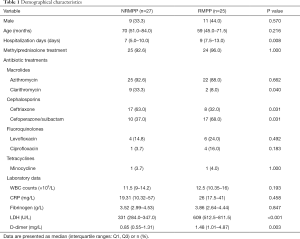
Full table
Comparison of microbial communities among the RMPP and NRMPP groups
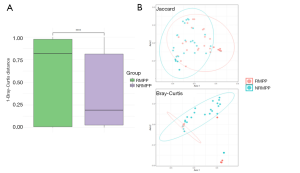
Principal coordinates analysis (PCoA) was used to compare microbial communities among the two groups. In addition, regarding beta diversity based on Bray-Curtis distance, there was a significant difference between the two groups (P<0.001, PERMANOVA), which means that the intra-group difference of RMPP group was smaller than that of NRMPP group (Figure 1A). PCoA plot based on neither Bray-Curtis distance nor Jaccard distance could effectively distinguish RMPP and NRMPP group (Figure 1B).
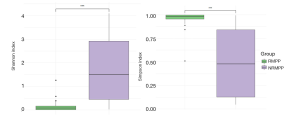
Alpha diversity analysis between the RMPP and NRMPP groups
Then alpha diversity was compared between the RMPP and NRMPP groups. Two of the alpha diversity calculated indexes, namely the Shannon and Simpson indices, representing the richness and evenness of the microbiota, were measured. A significant decrease in microbial diversity and increased dominance was observed in the samples collected from the RMPP group (Figure 2).
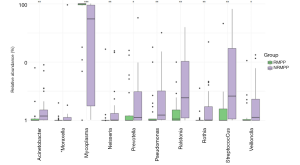
The most abundant taxa between the RMPP and NRMPP groups
We compared the most abundant phyla and genera between two groups (Figure 3; Table 2). At the phylum level, the top five phyla of the RMPP BALF microbiota were Tenericutes, Proteobacteria, Firmicutes, Bacteroidetes and Actinobacteria. As for the NRMPP group, the order of the top phyla was comparable to that of the RMPP group. Interestingly, the relative abundance of phylum Tenericutes significantly decreased in the NRMPP group whereas the Proteobacteria, Firmicutes and Actinobacteria were more abundant than RMPP group. At the genus level, the RMPP BALF microbiota was dominated by Mycoplasma, Streptococcus, Prevotella, Pseudomonas and Ralstonia. In the NRMPP group, the relative abundance of genus Mycoplasma was dramatically lower than the RMPP group. The relative abundance of commensal genera such as Strepcoccus, Neisseria, Veillonella, Rothia and Prevotella as well as that of the pathogenic genera, Acinetobacter, Pseudomonas and Ralstonia, was significantly higher in the NRMPP group (8). Additionally, we compared the M. pneumoniae load between the two groups. As shown in Table 2, M. pneumoniae load was significantly higher in almost all the RMPP patients than in the NRMPP group, indicating that M. pneumoniae occupied almost all of the niches in the LRT. In addition, we noticed that four patients from RMPP group had significantly lower M. pneumoniae relative abundance, and high load of human adenovirus 7 or human bocavirus type I was detected from these samples.
Full table
Biomarkers of the RMPP and NRMPP groups
LEfSe was used to explore the taxa with the strongest effect on different microbiology conditions among groups (Figure S1). This analysis detected two species from two genera as biomarkers for the RMPP group, and eleven species from seven genera for the NRMPP group. The Mycoplasma and Neisseria genera had the most powerful influence on RMPP, with Mycoplasma pneumoniae and Neisseria flavescens as the most abundant species. The most dominant NRMPP-associated biomarker were class Bacilli, genera Streptococcus, Ralstonia and species Ralstonia picketii and Streptococcus salivarius.
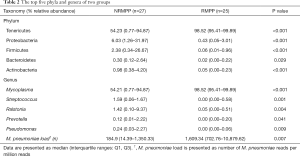
Differences in the distribution of antibiotic resistance genes (ARGs) and virulence factor genes (VFGs) between the RMPP and NRMPP groups
Reads from all samples were used to re-assemble the draft genome of M. pneumoniae, since data from a single group is not enough to obtain the complete genome (Figure S2). In total, 182,740 VFGs were uncovered in the M. pneumoniae genome including virulence-determined VFGs such as cytadherence accessory protein, community-acquired respiratory distress syndrome (CARDS) toxin, ADP1_MYCPN adhesin P1 and other virulence factors. The M. pneumoniae genome was found to consist of abundant ARGs against common antibiotics including fluoroquinolone, tetracycline, nitroimidazole, peptide, and macrolide. Next, we compared the annotated ARGs and VFGs in M. pneumoniae genome between NRMPP and RMPP groups (Figure 4). Interestingly, ARGs were successfully annotated in M. pneumoniae genome from 14 of 25 (56.0%) patients in RMPP group and 18 of 27 (66.7%) in NRMPP group. In addition, the NRMPP group held a greater number of annotated ARGs than the RMPP group as well as a higher relative abundance of macrolide ARGs (macB), fluoroquinolone resistance gene (patAB), nitroimidazole ARG (msbA) and peptide ARG (bcrA) (Figure 5A). It was suggested that the macrolide antibiotic resistance of M. pneumoniae in NRMPP may be higher than in RMPP. Moreover, the relative quantity of resistance genes of tetracycline antibiotics (tetT) in the RMPP group was higher than those of the NRMPP group. A 23rRNA gene single-nucleotide polymorphism mutation related to macrolide antibiotics resistance, A2063G, was detected from a patient from the RMPP group who received prolonged macrolide, cephalosporin, and fluoroquinolone antibiotics treatment (azithromycin, cefperazone-sulbactam and levofloxacin). Finally, The RMPP group had less annotated VFGs than NRMPP group while the relative abundance of adhesion-related virulence genes was higher (Figure 5B).

Microbial function differences between the RMPP and NRMPP groups
By means of LEfSe, we assessed the biomarker pathways of the BALF microbiota in children with RMPP (Figure S3). Microbial functions relating to metabolism of terpenoids and polyketides, lipid metabolism, DNA replication or repair and carbohydrate metabolism were found to be RMPP-associated pathways.
Association between BALF microbiota and hospitalization days
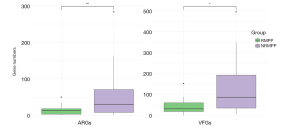
We used spearman correlation analysis to observe relationship between BALF microbiota and hospitalization days. In RMPP patients, higher relative abundance of Streptococcus oralis and Streptococcus pneumoniae as well as Streptococcus genera had a strong correlation with increased hospitalization days (Figure 6A). Interestingly, higher relative abundance of Streptococcus pneumoniae had a negative correlation with hospitalization days among NRMPP patients (Figure 6B). In addition, higher relative abundance of M. pneumoniae and lower Shannon index were also associated with increased hospitalization days in NRMPP group (Figure 6C,D).
Discussion
This is a pioneering study of LRT microbiome in children with RMPP. Although there have been many studies on the LRT microbiota of patients with pneumonia, the microbial function and host-microbial interaction remain to be explored, especially in children. It is clearly helpful to monitor changes in M. pneumoniae resistance and virulence, guide the use of antibiotics and, most importantly, advance our understanding of pathogenesis of RMPP.
The microbiota of LRT in children with RMPP is much simpler than that in children with NRMPP, and most of them are dominated by M. pneumoniae. Some researchers found that M. pneumoniae could eliminate other bacteria by directly competing for nutrients (9). Moreover, it clears other bacteria by activating the host inflammatory response (10). Together, these mechanisms could lead to decreased colonization of the commensal genera, Prevotella, Streptococcus, Veillonella and Rothia, in the respiratory tract of children with RMPP (8,11,12). In addition, the proliferation of pathogenic genera such as Pseudomonas and Acinetobacter were also inhibited. The microbial structure of RMPP group is highly homogeneous according to its lower intra-group beta diversity. Thus, such high similarity of microbial communities may be the signature of RMPP. In addition, the relative abundance of adherence-associated VFG was higher in RMPP group, as well as the more active KEGG pathway of glycolipid metabolism, which may be a key player in the virulence of M. pneumoniae (13). It is possible that a large number of M. pneumoniae can cause excessive inflammatory response and immune damage. M. pneumoniae secretes adhesion molecules with P1 adhesion protein to adhere to the epithelial cells, gradually reduces the function of phagocytes through fusion with host target cells, and causes immune damage (14). Simultaneously, M. pneumoniae infection stimulates the activation of T cells, and promotes higher expressions of cytokines and inflammatory factors, aggravating the inflammatory response (15-17). Such conditions may be due to the fact that the extensive use of broad-spectrum antibiotics such as cefoperazone in the RMPP group reduces the diversity of LRT, providing a less-competitive environment for M. pneumoniae to spread. Once M. pneumoniae occupies the whole niche, a large number of adhered airway epithelial cells become necrotic and exfoliate, inflammatory cells infiltrate and proinflammatory cytokines are secreted, and the release of CARDS toxin causes serious tissue damage to patients and such excessive activation of the immune system may be the main cause of RMPP (18).
Our results revealed that the NRMPP group were enriched for more ARGs than the RMPP group, and also showed a higher relative abundance of the macrolide ARG (macB), fluoroquinolone resistance gene (patA-B) and other ARGs (bcrA, msbA). Thus, the higher relative abundance of the macrolide ARG may not relate to RMPP. It was also reported that antibiotic-resistant M. pneumoniae infection often causes only mild pneumonia, while antibiotic-sensitive M. pneumoniae infection often causes RMPP, and M. pneumoniae resistance was not associated with serious treatment failure or the occurrence of serious complications, suggesting that the infection of resistant M. pneumoniae may not the main reason for RMPP (19). On the other hand, integrative and conjugative elements (ICEs) even the large chromosomal DNA fragments could be horizontally transferred among M. pneumoniae via conjugative transfers under the rigorous antibiotic selection pressure and complicated microbe-microbe interaction condition (20). Macrolides antibiotics, tetracycline and fluoroquinolones antibiotics are first-line and second-line drugs for the treatment of MPP, respectively. Such transfer of genetic components or ARGs (such as tetT, patA-B) may lead to widespread transfer of drug resistance genes among M. pneumoniae and the failure of treatment against MPP. In addition, many studies have suggested that point mutations in the 23S rRNA gene and the ribosomal proteins L4 and L22 are predominantly responsible for macrolide resistance in M. pneumoniae (21,22). Most of the mutations that found occurred at sites 2063 and 2064 in the 50 subunit 23S ribosomal RNA region V while some occurred at sites 2617 (C2617G, C2617A) (23,24). This gene mutation has been shown to induce macrolide resistance (PMCID: PMC529214) and also associated to restriction fragment length polymorphism in the P1 cytadhesion gene (PMCID: PMC228818). In our study, merely one patient who received prolonged and complex antibiotics treatment carried the A2063G mutation. Interestingly, macB, which was first identified as a macrolide antibiotic-specific transporter in E. coli, was widely identified in NRMPP patients (25). As a member of family of ATP-binding cassette-type transporters, it utilizes ATP hydrolysis as a driving force to pump out macrolide antibiotics. What should be recognized is that in China the macrolide-specific efflux system and modification of target sites are equally crucial mechanisms responsible for macrolide resistance in M. pneumoniae.
We uncovered that Streptococcus oralis and Streptococcus pneumoniae were associated with an increase in hospitalization days among RMPP patients. It is suggested that such species may be not conducive to the prognosis of RMPP. Actually, Streptococcus oralis was thought to be associated with ventilator-associated pneumonia and accelerated replication of influenza viruses in LRT while Streptococcus pneumoniae was a common pathogen of community-acquired pneumonia (26,27). Conversely, Streptococcus pneumoniae was negatively associated with prolonged hospitalization days in NRMPP group, while prolonged hospitalization days was associated with lower Shannon diversity and higher M. pneumoniae abundance. Thus, we proposed a hypothesis that the relative abundance and dominance of M. pneumoniae in LRT are factors determining the hospitalization days in the case of NRMPP due to its low density. For RMPP, the M. pneumoniae was close to covering the entire LRT, and the relative abundance of other pathogens became the determinant of the hospitalization days.
Although M. pneumoniae played an important role in RMPP pathogenesis, Neisseria flavescens was detected as a RMPP-associated biomarker via LEfSe. Neisseria flavescens is generally a commensal species but it turned out to be an important role in RMPP. In fact, Neisseria flavescens was previously reported to act as a pathogen in the LRT, and hence, it is possible that the pathogenic potential of such commensal species is activated in the inflammatory environment (28-30).
There are several limitations of this study. All children were treated with at least one antibiotic before bronchoscopy, and the treatments of ceftriaxone and cefoperazone/sulbactam were not balanced between groups, which may lead to temporary changes in the microecology of their LRT, as antibiotics themselves can reduce microbiota diversity. In addition, the number of patients in this study was not particularly large. An important limitation of mNGS is that it is only at the DNA level, and thus, it cannot distinguish between live and dead microorganisms and cannot determine the expression of related functional genes. Combined with metatranscriptomic sequencing, however, it can provide a real-life profile and function of microorganisms.
Conclusions
In summary, we explored the changes of microbiota structure and related functions in the LRT of patients with RMPP and NRMPP, which should advance our understanding of the pathogenesis of RMPP and provide foundations for further research.
Acknowledgments
Language polishing: Language was polished by the editing agency “EonBio” (Email: help@eonbio.com).
Funding: This work was supported by the National Natural Science Foundation of China (Grant code: 81601332).
Footnote
Reporting Checklist: The authors have completed the MDAR checklist. Available at http://dx.doi.org/10.21037/tp-20-404
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tp-20-404
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tp-20-404). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The ethical application and the consent procedure of this study were approved by the Ethics Committee of Children’s Hospital of Fudan University (No. 2016-87). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013), and informed consent was taken from all the parents.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kutty PK, Jain S, Taylor TH, et al. Mycoplasma pneumoniae Among Children Hospitalized With Community-acquired Pneumonia. Clin Infect Dis 2019;68:5-12. [PubMed]
- Lee H, Yun KW, Lee HJ, et al. Antimicrobial therapy of macrolide-resistant Mycoplasma pneumoniae pneumonia in children. Expert Rev Anti Infect Ther 2018;16:23-34. [Crossref] [PubMed]
- Yang TI, Chang TH, Lu CY, et al. Mycoplasma pneumoniae in pediatric patients: Do macrolide-resistance and/or delayed treatment matter? J Microbiol Immunol Infect 2019;52:329-35. [Crossref] [PubMed]
- Flanagan JL, Brodie EL, Weng L, et al. Loss of bacterial diversity during antibiotic treatment of intubated patients colonized with Pseudomonas aeruginosa. J Clin Microbiol 2007;45:1954-62. [Crossref] [PubMed]
- Shenoy MK, Iwai S, Lin DL, et al. Immune Response and Mortality Risk Relate to Distinct Lung Microbiomes in Patients with HIV and Pneumonia. Am J Respir Crit Care Med 2017;195:104-14. [Crossref] [PubMed]
- Subspecialty Group of Respiratory Diseases, The Society of Pediatrics, Chinese Medical Association; Editorial Board, Chinese Journal of Pediatrics. Guidelines for management of community acquired pneumonia in children (the revised edition of 2013) (I). Zhonghua Er Ke Za Zhi 2013;51:745-52.
- Lee WJ, Huang EY, Tsai CM, et al. Role of Serum Mycoplasma pneumoniae IgA, IgM, and IgG in the Diagnosis of Mycoplasma pneumoniae-Related Pneumonia in School-Age Children and Adolescents. Clin Vaccine Immunol 2017;24:e00471-16. [Crossref] [PubMed]
- Prevaes SM, de Steenhuijsen Piters WA, de Winter-de Groot KM, et al. Concordance between upper and lower airway microbiota in infants with cystic fibrosis. Eur Respir J 2017;49:1602235 [Crossref] [PubMed]
- Yang J, Hooper WC, Phillips DJ, et al. Cytokines in Mycoplasma pneumoniae infections. Cytokine Growth Factor Rev 2004;15:157-68. [Crossref] [PubMed]
- Peteranderl C, Sznajder JI, Herold S, et al. Inflammatory Responses Regulating Alveolar Ion Transport during Pulmonary Infections. Front Immunol 2017;8:446. [Crossref] [PubMed]
- Miller SI, Ernst RK, Bader MW. LPS, TLR4 and infectious disease diversity. Nat Rev Microbiol 2005;3:36-46. [Crossref] [PubMed]
- Zhao F, Liu G, Wu J, et al. Surveillance of macrolide-resistant Mycoplasma pneumoniae in Beijing, China, from 2008 to 2012. Antimicrob Agents Chemother 2013;57:1521-3. [Crossref] [PubMed]
- Blötz C, Stülke J. Glycerol metabolism and its implication in virulence in Mycoplasma. FEMS Microbiol Rev 2017;41:640-52. [Crossref] [PubMed]
- Krause DC, Leith DK, Wilson RM, et al. Identification of Mycoplasma pneumoniae proteins associated with hemadsorption and virulence. Infect Immun 1982;35:809-17. [Crossref] [PubMed]
- Chaudhry R, Varshney AK, Malhotra P. Adhesion proteins of Mycoplasma pneumoniae. Front Biosci 2007;12:690-9. [Crossref] [PubMed]
- Krause DC. Mycoplasma pneumoniae cytadherence: unravelling the tie that binds. Mol Microbiol 1996;20:247-53. [Crossref] [PubMed]
- Maselli DJ, Medina JL, Brooks EG, et al. The Immunopathologic Effects of Mycoplasma pneumoniae and Community-acquired Respiratory Distress Syndrome Toxin. A Primate Model. Am J Respir Cell Mol Biol 2018;58:253-60. [Crossref] [PubMed]
- Becker A, Kannan TR, Taylor AB, et al. Structure of CARDS toxin, a unique ADP-ribosylating and vacuolating cytotoxin from Mycoplasma pneumoniae. Proc Natl Acad Sci U S A 2015;112:5165-70. [Crossref] [PubMed]
- Narita M. Two unexpected phenomena in macrolide-resistant Mycoplasma pneumoniae infection in Japan and the unique biological characteristics of Mycoplasma pneumoniae. J Infect Chemother 2011;17:735-6. [Crossref] [PubMed]
- Citti C, Dordet-Frisoni E, Nouvel LX, et al. Horizontal Gene Transfers in Mycoplasmas (Mollicutes). Curr Issues Mol Biol 2018;29:3-22. [Crossref] [PubMed]
- Pereyre S, Goret J, Bébéar C. Mycoplasma pneumoniae: Current Knowledge on Macrolide Resistance and Treatment. Front Microbiol 2016;7:974. [Crossref] [PubMed]
- Bébéar C, Pereyre S, Peuchant O. Mycoplasma pneumoniae: susceptibility and resistance to antibiotics. Future Microbiol 2011;6:423-31. [Crossref] [PubMed]
- Copete AR, Aguilar YA, Rueda ZV, et al. Genotyping and macrolide resistance of Mycoplasma pneumoniae identified in children with community-acquired pneumonia in Medellín, Colombia. Int J Infect Dis 2018;66:113-20. [Crossref] [PubMed]
- Yang HJ, Song DJ, Shim JY. Mechanism of resistance acquisition and treatment of macrolide-resistant Mycoplasma pneumoniae pneumonia in children. Korean J Pediatr 2017;60:167-74. [Crossref] [PubMed]
- Kobayashi N, Nishino K, Hirata T, et al. Membrane topology of ABC-type macrolide antibiotic exporter MacB in Escherichia coli. FEBS Lett 2003;546:241-6. [Crossref] [PubMed]
- Nishioka K, Kyo M, Nakaya T, et al. Proteins produced by Streptococcus species in the lower respiratory tract can modify antiviral responses against influenza virus in respiratory epithelial cells. Microbes Infect 2021;23:104764 [Crossref] [PubMed]
- de Carvalho Baptista IM, Martinho FC, Nascimento GG, et al. Colonization of oropharynx and lower respiratory tract in critical patients: Risk of ventilator-associated pneumonia. Arch Oral Biol 2018;85:64-9. [Crossref] [PubMed]
- Huang L, Ma L, Fan K, et al. Necrotizing pneumonia and empyema caused by Neisseria flavescens infection. J Thorac Dis 2014;6:553-7. [PubMed]
- Johnson AP. The pathogenic potential of commensal species of Neisseria. J Clin Pathol 1983;36:213-23. [Crossref] [PubMed]
- Wu BG, Segal LN. The Lung Microbiome and Its Role in Pneumonia. Clin Chest Med 2018;39:677-89. [Crossref] [PubMed]


