Somatic DNA mutation analysis in targeted therapy of solid tumours
Cancer is a global health burden with 13-14 million new cases and 7.6-8.2 million cancer-related deaths per annum (1,2). Great effort and huge resources have been put into “the war on cancer” for the past four decades with the goals to cure cancer or to prolong life and to improve its quality. However, progress has been slow in the overall reduction of cancer mortality. For instance, lung cancer is the leading cause of cancer-related death worldwide (2,3) yet its 5-year survival remains 16.8% in the United States (4). This is essentially unchanged over the past two decades. Similarly, the median survival for patients with advanced melanoma is only 6-9 months (5,6).
The completion of the Human Genome Project in 2003 allowed the era of genomic medicine to start (7). The core goal of the Project was to utilise genomic knowledge for better treatment, prevention and so overall reduction in health costs. Cancer was shown to be a disease of the genome with different hallmarks including genome instability and an accumulation of somatic mutations (8,9). While cancers are characterised by numerous genomic aberrations, some acquired mutation(s) may be sufficient to induce growth and impaired differentiation leading to cancer development. This powerful somatic effect has been commonly described as a driver mutation and the overall phenomenon as oncogene addiction (9,10).
Oncogene addiction becomes the rationale for targeted therapy of solid tumours enabling a model that delivers treatment with a higher probability of efficacy while at the same time lowers the risk for adverse events (3,10). This biologically consistent approach saves time compared to a more trial-and-error strategy, improves the quality of life for cancer patients, and brings economic benefits by avoiding expensive but ineffective therapies. Targeted therapies for advanced lung cancer, melanoma, colorectal cancer and gastrointestinal stromal tumour (GIST) are examples of success stories that have resulted from the translation of knowledge gained from the Human Genome Project into informed choices in cancer treatments leading to prolonged survivals (7).
To facilitate the clinical applications of somatic DNA mutation analysis, we review the challenges involving current technologies and discuss the clinical implications of several actionable i.e., relevant to treatment, driver mutations in targeted therapy.
Somatic DNA mutation analysis
Clinicians and pathologists should be aware in the modern genomics-based therapeutic era that acquisition of tumour tissue in a biopsy is not just for histological diagnosis and staging, but increasingly for somatic DNA mutation analysis. Molecular characterisation is used to reveal underlying driver mutations and altered pathways which ultimately lead to more personalised or stratified targeted therapies (3).
Tumour DNA source
Somatic DNA mutation analysis requires tumour tissue to source DNA which should be extracted from a relatively pure population of tumour cells and without significant necrosis or inflammation. Surgical resection specimens are generally straightforward since tumour-rich regions are more easily located. However, small biopsies, a common source in practice, are more complex to use. There are different types of biopsies including: core biopsy, fine needle aspiration or cytology samples. The latter may be obtained through bronchoalveolar lavage, bronchial brushing, fine needle biopsy or the use of pleural/peritoneal fluids.
Small biopsies have inherent limitations since they represent a single snapshot in either time or space with the latter leading to selection bias due to heterogeneity. They also usually consist of an admixture of tumour and non-tumour cells, and require macrodissection to enrich for the tumour cell populations. Proper sampling and enrichment is needed to enhance the sensitivity and facilitate detection of low frequency mutations. Multiple sampling at primary and metastatic sites will help to overcome false negative results (11) but this option is not usually available.
Fresh tissue is ideal for somatic DNA testing because it delivers sufficient high quality DNA. While freezing has traditionally been used for storage, fresh tissue can also be kept in a preservative such as RNAlater (Life Technologies) for a convenient transit at room temperature. In reality, the most common source of diagnostic tumour DNA is from formalin-fixed paraffin embedded (FFPE) tissue which is the traditional method for preparing tissue sections for microscopic examination. It also allows verification of tumour material in the target region and macrodissection, if necessary. FFPE blocks are convenient for transport and long-term storage.
However, formalin fixation causes DNA fragmentation and cross-linkage and may introduce artefacts through stochastic deamination and/or depurination (12,13). In order to reduce these detrimental effects on DNA, tumour tissue should be fixed in 10% neutral buffered formalin under low temperature and without excessive length exposure (14). An alcohol fixative can be excellent for molecular characterisation since cytological smears or Diff Quik (DQ) samples often yield better quality DNA in our and other’s experience. Such material is air dried prior to alcohol fixation and does not require the use of formalin (15).
DNA assessment for reliable testing
Limited material remains a common obstacle for somatic DNA mutation analysis. An appropriate and validated DNA extraction method should be used to maximise the chance of obtaining suitable DNA from sparse FFPE tissue. It has been estimated that >1,000 tumour cells could be sufficient for analysis of common mutations in current clinical practice (15). In some cases with very limited material, even what comes from the needle rinse can provide useful material for analysis. Ultimately, the most sophisticated molecular techniques cannot compensate for insufficient and/or poor-quality material. Therefore, both quality and quantity of all target DNA templates should be carefully assessed before analysis.
Spectrophotometric analysis is commonly used for DNA assessment. High ratios of absorbance at 260/280 and 260/230 nm indicate samples are free from significant contaminations from protein, peptide and organic solvents. The quantity of DNA can be estimated based on the absorbance at 260 nm wavelength. However, this method may be inaccurate in FFPE samples since it can overestimate the quantity due to the presence of degraded DNA and RNA. Fluorometric measurement is a better method since it only detects double-stranded DNA. However, even double-stranded DNA may not equate to an amplifiable template due to excessive crosslink and fragmentation during formalin fixation. Various quantitative real time polymerase chain reaction (PCR) or mass spectrometric methods have been developed to assess the amount and amplification potential of FFPE DNA aiming at efficient use of available but limited DNA. Mass spectrometry analysis using the Exome ID kit (Agena Biosciences) allows different lengths and genomic loci to be assessed in a single assay and consumes only minute amount of DNA (16). Accurate DNA assessment is also essential for the next generation sequencing (NGS) analysis (17).
Challenges in somatic DNA mutation profiling
The ideal method for somatic DNA mutation analysis would: (I) identify a spectrum of genomic aberrations including single nucleotide variants, small indels, copy number aberrations and even structural variants such as re-arrangements or fusion genes; (II) work in a single test and without an excessive amount of DNA; (III) identify somatic DNA mutations in a background of normal with one or more subpopulations of tumour cells i.e., has sufficient sensitivity; (IV) utilise a comprehensive list of targeted genes to maximise the opportunity for targeted therapy; (V) be cost effective and deliver results in a timely fashion to facilitate early initiation of personalised treatment. No one assay currently meets all these criteria, and there remain a number of outstanding technologic challenges (Table 1).
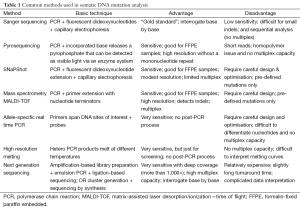
Full table
The first and foremost challenge is providing reliable and comprehensive results with limited tumour DNA. An international external quality assessment scheme has identified the major analytical methods to be Sanger sequencing and allele-specific real time PCR (Table 1) (18). Both amplify one DNA fragment at a time and thus consume significant amounts of DNA. Sequential analysis of various genes and regions with these approaches also limits timely delivery of results. While Sanger sequencing can interrogate every base and identify known and unknown variants, it involves many post-PCR manipulations such as clean-up of PCR and sequencing products. Its most critical disadvantage is the low sensitivity of this “gold standard”, which usually identifies a somatic DNA variant when present at 20% or more. Sanger sequencing can provide a false-negative result or no result in some cases (18-20).
The use of pyrosequencing significantly improves the sensitivity since detection is based on light emission after nucleotide incorporation (Table 1) (21,22). However, this method is limited by a very short sequencing read length (80-140 bp), similar post-PCR manipulations and no capacity for multiplexing.
The allele-specific real time PCR method eliminates post-PCR manipulations and is relatively simple to perform with superior sensitivity (1% detection limit) (20,22,23). However, allele-specific PCR can only identify predefined variants and requires careful design (due to limited primer options) and optimisation because of substantial risk of mispriming. Commercial kits are expensive, particularly if several are required to cover multiple genes. Allele-specific PCR is problematic when it comes to distinguishing different target nucleotides at the same position (20,23).
The accuracy of somatic DNA mutation analysis remains a concern as shown by only 72 of 91 participant laboratories passing an external quality assessment (18). Somatic DNA mutation analysis with multiplex capacity is advantageous since it can profile a multiple genes simultaneously while consuming acceptable amounts of DNA.
Extension-based assays such as SNaPShot® and mass spectrometry are commonly used for somatic DNA testing with acceptable sensitivity (Table 1) (24-26). Multiplex amplicons are generally around 80-120 bp, which makes the assay robust for fragmented DNA in FFPE samples. Extension-based assays are able to differentiate various nucleotides at the same position. The mass spectrometric assay has its own unique features including: (I) high resolution without nucleotide labelling or modification; (II) high multiplex capacity (up to 50 targets per reaction); (III) additional specificity related to the extension probe in specific and high multiplex PCRs (24-26). Disadvantages are primer extension assays can only detect pre-defined variants, and undefined ones will be missed. Parallel fluorescence in situ hybridisation (FISH) is required for the detection of cancer gene amplifications, rearrangements or gene fusions (27).
A second challenge is distinguishing false-negative results from tumour heterogeneity (11), amplification failure or high background noise. False negative results can be as high as 6.7-7.6% (18) and may deprive patients of the opportunity for targeted therapy. A common cause for this type of result is a low proportion of tumour cells. Therefore, it is important to have the slides reviewed by a tissue pathologist before molecular testing. Working with insufficient DNA templates is not uncommon due to severe fragmentation and cross-linkage even if spectrophotometric or fluorometric values suggest that a defined quantity of DNA is present. Extracted DNA that has contaminants can also lead to false-negative results. For example, melanin is frequently present in melanoma samples and requires additional clean-up to remove this PCR inhibitor.
Many allele-specific PCR assays utilise a template control in addition to the target amplicon to identify when DNA has failed to be added. However, amplification of non-target control does not necessarily mean that target template is available for amplification. Extension-based assays are preferred since they have a built-in template control and any amplicon will be interrogated by a designated extension primer irrespective of the presence or absence of a predefined mutation. Random somatic DNA variants represent another source for amplification failure if they occur at the primer binding sites. Using a set of redundant primers is helpful in all PCR-based somatic DNA tests since they reduce the interference problem due to potential binding site variants.
The third and last challenge is how to avoid false positives found in 0.6-1.4% of molecular assays (18). False-positive findings can be problematic because: (I) they lead to unnecessary treatment with what are likely to be expensive drugs; (II) the patient will be exposed to side effects associated with a drug that is unlikely to work. Treatment with the wrong drug can even accelerate tumour growth as exemplified by giving a patient with melanoma a BRAF inhibitor based on a false-positive BRAF p.V600E mutation result when the tumour actually carries a RAS mutation (28-30); (III) patients treated inappropriately could miss out on the optimal time for effective treatment.
As discussed above, formalin fixation can introduce artefactual changes from cytosine to uracil (becoming thymine in PCR product) due to deamination or single base deletion secondary to depurination (12,13). Such artefacts appear to be “convincing” after amplification, particularly when there is insufficient DNA. PCR amplified artefacts will not be prominent if there are sufficient DNA templates since deamination or depurination are stochastic events during formalin fixation. It is also possible to excise the introduced uracil through the uracil-DNA glycosylase digestion (13,31) provided there is sufficient amount of DNA left for somatic mutation analysis.
Role of next generation sequencing (NGS) in somatic DNA mutation analysis
NGS or massively parallel sequencing as it is also called is a promising technology for somatic DNA mutation analysis. FFPE DNA can be amplified directly or after hybrid capture in a highly multiplexed fashion. This amplification strategy overcomes the issue of limited tumour DNA and so to some extent avoids the direct use of poor quality FFPE DNA as a sequencing template. NGS can interrogate every base in particular gene targets at an acceptable coverage (>1,000×) to ensure accuracy and sensitivity (17,22). It automatically controls the target amplification. NGS has significantly high throughput and can cover the long tail pattern for mutation distribution in many solid tumours. It can readily identify single nucleotide variants and small indels, and potentially copy number aberrations and rearrangement or fusions in a single assay. NGS saves precious tumour tissue, avoids time-consuming sequential testing, and so maximises the patient’s opportunity for targeted therapy. Further improvement can be achieved if redundant hybridisation probes or amplification primers can be introduced to prevent allele dropout(s) due to potential primer binding site variants.
Nevertheless, there are disadvantages with NGS when it is used for clinical diagnosis: (I) the turnaround time will be longer; (II) it is relatively expensive considering the need for specialised equipment, maintenance, sequencing costs and computer infrastructure for analysis and data storage (17); and (III) more effort and bioinformatics skills are required for data analysis. Whole exome NGS lacks the potential to detect rearrangements or fusions, while whole genome NGS is not suitable for high coverage rates with current costs. At present, these problems will make it difficult to use the NGS applications of whole exome or genome sequencing. If NGS is used, it will be targeted to particular gene(s).
Implications of molecular characterisation
In the past decade, extensive research into molecular genetics of lung cancer has identified different driver mutations, deciphered the underlying pathways involved in pathogenesis, and from this has emerged the concept of targeted therapy (3,14,32-34). The translation of this knowledge into clinical practice has changed the management of advanced non-small cell lung carcinoma (NSCLC) and altered its nature course. It has led to a paradigm shift in clinical oncology from organ- and morphology-based to gene-based practice. The detection of somatic DNA involving solid tumours is increasingly playing a key role by allowing more biologically relevant diagnoses to be made leading to more effective therapies to be selected (3). The same applies to detection of somatic cell based RNA and chromosomal aberrations although these are not the subject of this review.
Somatic DNA mutations in the tyrosine kinase receptors
EGFR—epidermal growth factor receptor
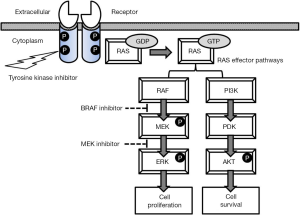
EGFR encodes a transmembrane receptor tyrosine kinase (Figure 1) and binds several ligands including the epidermal growth factor, transforming growth factor-alpha and amphiregulin. In responding to the ligand binding, the receptor forms a homodimer or heterodimer, followed by autophosphorylation in the activation loop of the catalytic tyrosine kinase domain. The active (autophosphorylated) kinase state controls downstream MAPK, PI3K and STATs pathways, regulating cell proliferation and enhancing cell survival (Figure 1) (34).
In NSCLC, somatic DNA mutations can occur in or close to the EGFR kinase domain (exons 18 to 21) (18) and activate kinase activity by abrogating autoinhibition. The most abundant mutations are small in-frame deletions in exon 19 encompassing the leucine-arginine-glutamate-alanine motive (45-50% of mutations) and the single nucleotide missense variant p.L858R (40-45%) (24,32,33). Exon 19 deletions remove residues from the activation loop and structurally impair the ability of the protein to adopt its inactive position (35-37). The p.L858R variant mutation occurs within the activation loop and leads to a shift in the kinase towards an activated state (36). Both mutation classes result in a decreased affinity for ATP, but enhanced affinity for the tyrosine kinase inhibitors (TKIs, discussed below) compared with the wild-type receptor (35-37). They represent the classical activating mutations and have oncogenic capability for transforming fibroblast and lung epithelial cells (38,39). Interestingly, activating mutations are observed in 10-15% of NSCLC in western Europeans but up to 25-30% in East Asians, particularly female non-smokers.
Currently there are two classes of EGFR antagonists: (I) anti-EGFR monoclonal antibodies such as cetuximab and panitumumab (40). They bind specifically to the extracellular domain of the receptor and block ligand binding, thus preventing ligand-induced EGFR activation. These antibodies also promote receptor internalisation and antibody or complement-mediated cytotoxicity; (II) small-molecule ATP-mimetic TKIs bind to the intracellular catalytic domain of the receptor to inhibit EGFR tyrosine phosphorylation and downstream signalling pathways (Figure 1). The response rates following treatment with TKIs is >58-70%, with median progression-free survival (PFS) >9 months and overall survival (OS) of 24-30 months in NSCLC patients with activating mutations (41,42). TKIs have become the most robust initial therapy for these patients (43). Other mutations including p.G719A/C/S and p.L861Q (<5%) are also associated with some TKI sensitivity although there are less data about responses in these rare mutations (44).
Several EGFR mutations produce resistance to targeted therapy. These include the missense mutation p.T790M, small insertions/duplications of exon 20 and missense mutations at p.S768 and p.V769 (37,39,45). p.T790M at the gatekeeper position of the ATP kinase pocket can mitigate the sensitisation effect of activating mutations (36). The underlying mechanism is due to an increase in the receptor affinity for ATP, and disruption of kinase-drug binding (36,38). Exon 20 indels (5% of mutations) disturb the structural orientation that controls ATP and TKI binding (36), affect the TKI affinity to the receptor and promote the active state of the kinase domain.
The mutation p.T790M is also the major change (50-60%) involved in acquired TKI resistance (37,45,46). TKI Afatinib has some effect on p.T790M-related resistance, but the third-generation TKIs may be even more potent (3,47). Acquired resistance can result from the bypassing of classical signalling pathways (around <15%) involving MET (46,48), ERBB2 (49) and others (47,50) although they are individually uncommon and can be co-identified with p.T790M in same specimens.
KIT—V-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog
KIT is a member of the type 3 transmembrane receptor tyrosine kinase family and encodes the human homolog of the proto-oncogene c-kit. Stem cell factor is its ligand. KIT has similar response to ligand binding as EGFR, i.e., receptor dimerization, autophosphorylation, receptor activation (51,52), and it regulates cell proliferation and survival (Figure 1).
KIT activating mutations are mostly found in the juxtamembrane exon 11 (65% of mutations) and include in-frame deletions (e.g., around p.W557 and p.K558), missense mutations (e.g., p.L576P) or complex changes. As derived from analog studies (in silico analysis of tyrosine receptor analogs allowing inference from known protein functions), the juxtamembrane domain could act as a negative regulator of kinase. These mutations could disrupt KIT conformational integrity and impair its regulation. Other less common clusters are on exon 9 (10%) and exon 13 (2%) (53). KIT exon 9 codes for the extracellular domain (the 5th immunoglobulin-like loop) and the mutations in exon 9 can activate the kinase in the absence of ligand via the stabilisation of receptor dimers (54). KIT activating mutations are oncogenic with a gain-of-function and have been found in 80-85% of adult GIST as well as chronic myeloid leukaemia (55), seminoma and melanoma (56-58).
Imatinib was initially developed to treat BCR-ABL chronic myeloid leukaemia (59) and has been successfully used in GIST and melanoma patients with KIT mutations. Imatinib selectively inhibits tyrosine kinase including ABL, BCR-ABL, KIT and PDGFR (60). It reversely binds to the ATP binding pocket of the KIT or ABL kinase and locks it in a self-inhibited conformation (61). This process inhibits kinase activity, switches off the downstream signalling pathways, leads to growth arrest and eventual apoptosis in tumour cells (62). Imatinib can achieve disease control in 70-85% of patients with KIT mutations with a median PFS of 20-24 months, and an estimated OS >36 months (63,64). TKI therapy has become the standard of care for patients with advanced GIST. Therapy is continued while patients are experiencing clinical benefit. Patients with exon 11 missense changes or insertions have a favourable prognosis, whereas those with KIT exon 11 p.W557-K558del and/or exon 9 mutations have a poor prognosis (65).
While TKI therapy induces GIST regression, imatinib rarely achieves complete remissions. Even long-term TKI therapy fails to eradicate tumour cells. Most patients who respond will eventually develop acquired resistance. Antonescu et al. reported that the 2-year survival of the imatinib-treated patients with advanced GIST was as high as 72%, but in half the disease progressed within two years (66). Innate resistance occurs in 10-15% of GIST patients. Patients with KIT exon 11 or 9 exon mutations or wild-type GIST have a 5%, 16% and 23% probability of demonstrating innate imatinib resistance (67).
Acquired resistance to imatinib commonly occurs through the emergence of second-site mutations in cis with the original KIT mutations (9,66,68,69). These mutations are mainly clustered in either the ATP binding pocket (p.V654A and p.670I) or the kinase activation loop (p.C809G, p.D816H, p.D820G/A, p.N822K/Y and p.Y823D) (9,70). The mutations bypass the inhibitory effects of the drug by interference with imatinib binding or direct activation (66). In the minority GIST patients, other mechanisms may be involved such as KIT genomic amplification or activation of an alternative tyrosine kinase receptor (71).
Somatic DNA Mutations in RAS proteins
RAS proteins are small GTPases that cycle between an active (GTP) or an inactive GDP bound state (Figure 1). There are three human RAS genes encoding highly homologous 21 kDa proteins. KRAS and HRAS were first identified in the Kirsten and Harvey strains of mouse sarcoma virus, whereas NRAS represents the neuroblastoma RAS viral oncogene homologue. RAS proteins link the activation of cell surface receptors with a wide variety of cellular processes leading to the control of proliferation, differentiation and apoptosis (Figure 1). RAS activation can result from somatic DNA mutations, upstream activation of tyrosine kinase receptors or by loss of function of regulating tumour suppressor genes. Furthermore, oncogenic RAS proteins can interfere with metabolism of tumour cells, microenvironment remodelling, evasion of immune response, and can contribute to the metastatic process. Efforts to target RAS mutants directly have thus far been unsuccessful.
KRAS—Kirsten rat sarcoma viral oncogene homolog
KRAS missense mutations at codon 12 or 13 are most common and account for approximately 85% of somatic DNA mutations in colorectal cancer. Other activating mutations at codons 61, 117 or 146 are found in up to 15% of KRAS mutant cases (72-74). The replacement of p.G12 is associated with steric hindrance of GTPase-mediated GTP hydrolysis and thus promotes the formation of constitutive activation. Glutamine substitution at codon 61 impairs GTPase activity by disrupting a hydrogen bond with GAPp120 (75). KRAS p.A146 mutations do not impair KRAS GTPase activity, but confer activity by increasing the rate of guanine nucleotide exchange (76). Their effect may be augmented by frequent conversion to homozygosity and low-level copy number gain of the KRAS gene locus (73). Both p.K117 and p.A146 missense mutations are associated with relatively lower levels of GTP-bound RAS compared to the p.G12D mutant and usually predict a more favourable clinical outcome (73,74). KRAS somatic DNA mutations have been identified in a variety of human malignancies, most frequently in pancreatic cancer, NSCLC and colorectal cancer.
The efficacy of anti-EGFR monoclonal antibodies is confined to patients with wild-type KRAS in metastatic colorectal patients. Therefore, KRAS mutation testing is critical before starting targeted therapy. Patients with KRAS mutations, particularly in codon 12 or 13 should not receive this therapy (75). More recently, mutations such as changes at p.K117 and p.A146 in KRAS exon 4 have been shown to predict a lack of benefit from anti-EGFR antibody therapy (74). KRAS mutations are associated in 35-45% of cases with resistance to anti-EGFR monoclonal antibody.
In NSCLC, substitutions at KRAS codons 12 or 13 are more common (>97% of KRAS mutations). They are more likely to be found in lung adenocarcinomas from former or current smokers although they are also present in around 15% of never-smokers (77). It has been reported that NSCLC with KRAS mutations is more likely to present with locally advanced disease and a poorer survival rate (78). KRAS-mutant lung tumours are resistant to EGFR TKIs (79) although KRAS testing has not been widely adopted in non-multigene testing in NSCLC.
NRAS—neuroblastoma viral (V-Ras) oncogene homolog
Somatic NDA mutations of NRAS are commonly encountered in codons 12, 13 and 61. These mutations lock NRAS into a constitutively activated state by eliciting downstream effectors. Activating NRAS mutations are reported in 15-20% of advanced melanoma in Caucasian patients (80) and our unpublished data suggest that the prevalence of NRAS mutations can be up to 30% in the same ethnic group. In African and Asian populations, there is a lower frequency (12% and 7.2%, respectively) (81,82). Many studies have suggested that NRAS mutations are significantly more common in melanomas arising in chronic sun-damaged skin (83,84).
Newly emerged NRAS mutations that arise during treatment, such as p.Q61K, represent one of the resistance mechanisms for BRAF inhibitor therapy (85). MEK is downstream of BRAF in the MAPK pathway (Figure 1) and its inhibitor can mediate blockade of NRAS mutant signalling. MEK162 is an oral MEK inhibitor, which was tested in patients with advanced melanoma harbouring NRAS mutations. The results were encouraging, but the response rate was relatively low (<20%) (86). Most of the patients rapidly develop resistance to the MEK inhibitor. More trials are on the way to test the efficacy of new MEK inhibitors. The results so far suggest that single-agent strategies may prove insufficient in NRAS mutant tumours. Instead, combination strategies using a BRAF inhibitor with a MEK or AKT inhibitor may work synergistically to inhibit proliferation of tumour and resistant cells to overcome resistance. Interestingly, NRAS mutations in advanced melanoma can be a biomarker for response to immunotherapy since more clinical benefit was observed in those patients with NRAS mutants compared to those with RAF/NRAS wild types (87,88). Lung cancers harbouring NRAS mutations are a distinct subset with potential sensitivity to MEK inhibitors (89).
Somatic DNA Mutations in Serine/threonine Kinase
BRAF—v-raf murine sarcoma viral oncogene homolog B1
BRAF encodes a serine/threonine kinase with the monomer representing the “off” state. BRAF can be phosphorylated at p.T599 and/or p.S602 by RAS, followed by dimerization with itself or ARAF and CRAF. RAF dimer is the “on” state and can transmit proliferative and survival signals downstream of the RAS proteins (Figure 1).
BRAF p.V600E represents a hot-spot for mutations accounting for at least 75% of driver mutations in this gene, followed by p.V600K (20%) and others. Position 600 and its vicinity are part of the activation loop of the kinase. The replacement of a negatively charged glutamic acid to valine can disrupt the domain conformation and mimic the conformation of the phosphorylated wild-type protein, which is necessary for kinase activation (90). It can dramatically increase BRAF activity and lead to constitutive ERK activation (Figure 1) (91). Recently, a crystal structure study has revealed that dimerization is a key step for RAF activation. The p.V600E missense substitution significantly contributes to the destabilisation of the “off” conformation and the stabilisation of the “on” conformation through salt-bridge interactions (92). BRAF somatic DNA mutations have been found in 50% patients with advanced melanoma as well as 10% of colorectal cancer, 40% papillary thyroid cancer and others (93-95).
The outlook for advanced melanoma has been transformed with BRAF inhibitors including vemurafenib, dabrafenib and LGX818 in the past a few years (88). These inhibitors are selective ATP-competitors and can stabilise BRAF mutants in the ATP pocket (96). Vemurafenib and dabrafenib have a response rate of 48% compared to 5% with standard chemotherapy (6,85,97). The median OS increased to 13-16 months, but PFS was only 5-7 months. The limited data have suggested that low-activity BRAF mutations such as p.L597S can be suppressed by MEK inhibitor both in vitro and in melanoma patient (98).
Early development of resistance is the major drawback of BRAF inhibition therapy. Resistance can be attributed to several factors including induction of alternative splice variants of BRAF or de novo mutations in NRAS or MEK. BRAF splice variants lack the RAS-binding domain, but retain RAF kinase activity in the presence of vemurafenib secondary to their enhanced homodimerisaton (99,100). Upregulation of signalling through receptor tyrosine kinase in alternative proliferative pathways is also associated with both innate and acquired resistance (101). Inhibited BRAF can still activate the pathway through dimerization with CRAF (29,102).
A complete inhibition of the MAPK pathway can be achieved by the combination of BRAF and MEK inhibitors (Figure 1) (87). Combination treatment can delay or prevent MAPK-dependent resistance and reduce BRAF inhibitor related toxicities as a result of paradoxical activation of the MAPK pathway in non-melanoma BRAF wild-type cells (28,87). This strategy has proved effective and reduces the incidence of secondary malignancies arising from off target promotion of RAS mutant cancers such as squamous cell carcinoma (28). The combination of dabrafenib with trametinib in BRAF “addicted” cancers showed a significant higher response rate (76% vs. 54%) and significantly longer PFS (9.4 months versus 5.8 months) than monotherapy with dabrafenib (103). It has been proposed that this type of combination therapy along with immunotherapies is likely to replace BRAF inhibitor monotherapy as the preferred first-line MAPK inhibitor treatment for BRAF-mutant metastatic melanoma in the near future (3,88).
Somatic DNA mutations identified through multigene analysis
Multigene profiling can provide useful and, at times, unexpected information for clinical decision making. Somatic DNA mutations in EGFR, KRAS or NRAS and BRAF are generally mutually exclusive. Identification of one gene mutation provides confidence in a negative result for mutations in other genes. Patients with melanoma or GIST can benefit from simultaneous profiling of BRAF and KIT. Although rare, KIT mutations are found in melanomas from acral melanoma, mucosal melanoma and melanoma located in sun-damaged skin (57,104). It has also been reported that 15% of anal melanomas harboured a KIT mutation (104).
KIT TKIs (imatinib and sunitinib) are of interest in terms of melanoma treatment. Sunitinib has shown a clinical response in three of four KIT-mutated melanomas, but only in one of six melanomas with KIT amplification only (105). Melanoma patients with KIT mutation had a better outcome after imatinib treatment compared with those having BRAF inhibitor treatment. On the other hand, a primary BRAF mutation can be found in 7-13% of adult GIST patients who lack KIT/PDGFRA mutations (106). BRAF mutations can also be associated with acquired resistance when KIT-dependant GIST is treated with imatinib (107).
As discussed in the section on RAS proteins, the presence of wild-type KRAS is required but not sufficient to confer sensitivity to anti-EGFR monoclonal antibodies. The expression levels of EGFR ligands, increased EGFR copy number, NRAS or BRAF mutations and PTEN loss may contribute to non-responsiveness with therapy. Wild type status in the above genes is associated with an improved objective response, longer median PFS and OS. The finding of BRAF mutations in colon cancer can have different implications, for example, there may be no role for anti-EGFR treatment because of a rapid feedback activation mechanism (108). The inhibitory effect of the driver mutation p.V600E can evoke a rapid feedback activation of EGFR and support continued proliferation (108,109).
The activation of multiple pathways may be complementary and interchangeable across different cancers. BRAF mutation can trigger resistance to TKIs in EGFR-mutant lung cancer (110), while EGFR mutation can mediate resistance to vemurafenib in colon cancers with the BRAF p.V600E mutation (111). Newly emerged oncogenic RAS mutation has been shown to have high risk for the development of secondary malignancies in patients with selective BRAF inhibition therapy (28,112). Therefore, it is important to use a multigene analysis approach to monitor the underlying molecular changes at different stages during cancer treatment.
Conclusions and future prospects
Current insight into the cancer genome, particularly the identification of driver mutations, has invigorated the campaign against cancer. The stunning initial success in personalised targeted therapy has boosted optimism that cancer can be cured, and success in genomic medicine will continue to gain momentum. Genomic application will extend to paediatric malignancies, particularly to sarcomas (113). NGS screening of enriched targeted transcripts can be developed for more effective and accurate detection of a variety of driver fusion genes (114). Fusion gene identification will help the diagnosis and provide the prognostic information. Ultimately, it may lead to effective targeted therapies such as Crizotinib or Ceritinib for ALK-EML4 fusion gene in NSCLC (115). Cancer somatic DNA mutation analysis is crucial for precision oncology and tomorrow’s cancer treatment will be more dependent on mutations in a tumour than on the organ in which the cancer arises. Further research involving single tumour cell analysis might provide insights into inter- and intra-patient tumour heterogeneity, and tumour genomic, genetic and epigenetic evolution (3), which will help to overcome resistance to current targeted therapies.
Multiple sampling of blood from a cancer patient is described as a “liquid biopsy”. It allows circulating cell-free DNA derived from tumour to be monitored and analysed before, during and after the treatment (3,22). Tumour burden, residual disease, resistance and early relapse can be objectively detected during treatment. This level of molecular information will assist oncologists to be proactive rather than reactive once resistance or relapse is first detected.
Today, the bottleneck in cancer DNA mutation profiling is less related to data generation but revolves around data analysis, display and integration. More innovative approaches are essential, for example identifying and excluding formalin induced artefacts, detecting low frequency mutations and differentiation of driver mutations from a sea of passenger variants. We need the capability to visualise the vast data sets, identify hidden patterns and potential “Achilles’ heel” for particular cancers (10) as well as retrieve the relevant data for pharmacokinetic and pharmacodynamic consideration.
Data integration is essential in a multidimensional way to allow a “Google map” view for ease of use and interpretation. From such a map one can find not only the location of a target, but also its function and matched intervention(s). Contemporary knowledge from a variety of databases needs to be integrated in ways that allow better data interpretation, and to assist in decision making for individual cancer patients (3). As the numbers of tumour genome profiles grow, it is inevitable that new targets for treatment will be detected. Ultimately, improvements in patient outcomes will follow.
Acknowledgements
None.
Footnote
Conflicts of Interest: BY and RJT declare no conflict of interest. SOT has received honoraria from Roche, Pfizer and Astra Zeneca.
References
- Stewart B, Wild CP. International Agency for Research on Cancer, WHO. (2014) World Cancer Report 2014. Available online: http://www.thehealthwell.info/node/725845 (Accessed on 8th April 2015).
- Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin 2011;61:69-90. [PubMed]
- Masters GA, Krilov L, Bailey HH, et al. Clinical cancer advances 2015: annual report on progress against cancer from the american society of clinical oncology. J Clin Oncol 2015;33:786-809. [PubMed]
- Howlader N, Noone AM, Krapcho M, et al. eds. SEER Cancer Statistics Review (CSR) 1975-2011. Bethesda: National Cancer Institute, 2013.
- Balch CM, Gershenwald JE, Soong SJ, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol 2009;27:6199-206. [PubMed]
- Tronnier M, Mitteldorf C. Treating advanced melanoma: current insights and opportunities. Cancer Manag Res 2014;6:349-56. [PubMed]
- Green ED, Guyer MS. National Human Genome Research Institute. Charting a course for genomic medicine from base pairs to bedside. Nature 2011;470:204-13. [PubMed]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [PubMed]
- Macconaill LE, Garraway LA. Clinical implications of the cancer genome. J Clin Oncol 2010;28:5219-28. [PubMed]
- Weinstein IB, Joe AK. Mechanisms of disease: Oncogene addiction--a rationale for molecular targeting in cancer therapy. Nat Clin Pract Oncol 2006;3:448-57. [PubMed]
- Saint-Jean M, Quéreux G, Nguyen JM, et al. Is a single BRAF wild-type test sufficient to exclude melanoma patients from vemurafenib therapy? J Invest Dermatol 2014;134:1468-70. [PubMed]
- Lillie RD, Pizzolato P, Henderson R, et al. The influence of formaldehyde fixation time on the rate of nitrous acid deamination of erythrocytes and spinal cord proteins. Histochemie 1970;24:156-8. [PubMed]
- Do H, Dobrovic A. Dramatic reduction of sequence artefacts from DNA isolated from formalin-fixed cancer biopsies by treatment with uracil- DNA glycosylase. Oncotarget 2012;3:546-58. [PubMed]
- Lindeman NI, Cagle PT, Beasley MB, et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J Mol Diagn 2013;15:415-53. [PubMed]
- Lewandowska MA, Jóźwicki W, Jochymski C, et al. Application of PCR methods to evaluate EGFR, KRAS and BRAF mutations in a small number of tumor cells in cytological material from lung cancer patients. Oncol Rep 2013;30:1045-52. [PubMed]
- Irwin D, Hunt P, Pearce M. Exome ID ABS panel application note. Available online: http://agenabiocom/sites/default/files/41-20016R10-Exome-ID-AbS-Application-Note-031914pdf: Agena Biosciences, 2014.
- Yu B. Setting up next-generation sequencing in the medical laboratory. Methods Mol Biol 2014;1168:195-206. [PubMed]
- Patton S, Normanno N, Blackhall F, et al. Assessing standardization of molecular testing for non-small-cell lung cancer: results of a worldwide external quality assessment (EQA) scheme for EGFR mutation testing. Br J Cancer 2014;111:413-20. [PubMed]
- Luquin N, Yu B, Trent RJ, et al. DHPLC can be used to detect low-level mutations in amyotrophic lateral sclerosis. Amyotroph Lateral Scler 2010;11:76-82. [PubMed]
- Anderson S, Bloom KJ, Vallera DU, et al. Multisite analytic performance studies of a real-time polymerase chain reaction assay for the detection of BRAF V600E mutations in formalin-fixed, paraffin-embedded tissue specimens of malignant melanoma. Arch Pathol Lab Med 2012;136:1385-91. [PubMed]
- Ronaghi M, Uhlén M, Nyrén P. A sequencing method based on real-time pyrophosphate. Science 1998;281:363,365.
- Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86. [PubMed]
- Wenceslao SM, Gale J, Vasef MA. Detection of BRAF V600K mutations in melanoma: a comparison of Cobas 4800 BRAF V600 mutation test and laboratory developed pyrosequencing assay. J Mol Diagn 2012;14:637-748 (abstract ST16).
- Yip PY, Yu B, Cooper WA, et al. Patterns of DNA mutations and ALK rearrangement in resected node negative lung adenocarcinoma. J Thorac Oncol 2013;8:408-14. [PubMed]
- Su Z, Dias-Santagata D, Duke M, et al. A platform for rapid detection of multiple oncogenic mutations with relevance to targeted therapy in non-small-cell lung cancer. J Mol Diagn 2011;13:74-84. [PubMed]
- Thomas RK, Baker AC, Debiasi RM, et al. High-throughput oncogene mutation profiling in human cancer. Nat Genet 2007;39:347-51. [PubMed]
- O'Toole S, Yu B, Cooper W. Molecular Diagnosis in Cytology and Its Place in the New Classification: A Practical Guide. J OncoPathol 2014;2:63-73.
- Su F, Viros A, Milagre C, et al. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N Engl J Med 2012;366:207-15. [PubMed]
- Gibney GT, Messina JL, Fedorenko IV, et al. Paradoxical oncogenesis--the long-term effects of BRAF inhibition in melanoma. Nat Rev Clin Oncol 2013;10:390-9. [PubMed]
- Sanchez-Laorden B, Viros A, Girotti MR, et al. BRAF inhibitors induce metastasis in RAS mutant or inhibitor-resistant melanoma cells by reactivating MEK and ERK signaling. Sci Signal 2014;7:ra30. [PubMed]
- Pearl LH. Structure and function in the uracil-DNA glycosylase superfamily. Mutat Res 2000;460:165-81. [PubMed]
- Jänne PA, Engelman JA, Johnson BE. Epidermal growth factor receptor mutations in non-small-cell lung cancer: implications for treatment and tumor biology. J Clin Oncol 2005;23:3227-34. [PubMed]
- Pao W, Miller VA. Epidermal growth factor receptor mutations, small-molecule kinase inhibitors, and non-small-cell lung cancer: current knowledge and future directions. J Clin Oncol 2005;23:2556-68. [PubMed]
- Sharma SV, Bell DW, Settleman J, et al. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 2007;7:169-81. [PubMed]
- Kumar A, Petri ET, Halmos B, et al. Structure and clinical relevance of the epidermal growth factor receptor in human cancer. J Clin Oncol 2008;26:1742-51. [PubMed]
- Eck MJ, Yun CH. Structural and mechanistic underpinnings of the differential drug sensitivity of EGFR mutations in non-small cell lung cancer. Biochim Biophys Acta 2010;1804:559-66.
- Mulloy R, Ferrand A, Kim Y, et al. Epidermal growth factor receptor mutants from human lung cancers exhibit enhanced catalytic activity and increased sensitivity to gefitinib. Cancer Res 2007;67:2325-30. [PubMed]
- Yun CH, Mengwasser KE, Toms AV, et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A 2008;105:2070-5. [PubMed]
- Greulich H, Chen TH, Feng W, et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. PLoS Med 2005;2:e313. [PubMed]
- Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med 2008;358:1160-74. [PubMed]
- Costa DB, Kobayashi S, Tenen DG, et al. Pooled analysis of the prospective trials of gefitinib monotherapy for EGFR-mutant non-small cell lung cancers. Lung Cancer 2007;58:95-103. [PubMed]
- Maemondo M, Inoue A, Kobayashi K, et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med 2010;362:2380-8. [PubMed]
- Ettinger DS, Wood DE, Akerley W, et al. Non-small cell lung cancer, version 1.2015. J Natl Compr Canc Netw 2014;12:1738-61. [PubMed]
- Yeh P, Chen H, Andrews J, et al. DNA-Mutation Inventory to Refine and Enhance Cancer Treatment (DIRECT): a catalog of clinically relevant cancer mutations to enable genome-directed anticancer therapy. Clin Cancer Res 2013;19:1894-901. [PubMed]
- Engelman JA, Mukohara T, Zejnullahu K, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest 2006;116:2695-706. [PubMed]
- Engelman JA, Mukohara T, Zejnullahu K, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest 2006;116:2695-706. [PubMed]
- Chong CR, Jänne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat Med 2013;19:1389-400. [PubMed]
- Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007;316:1039-43. [PubMed]
- Takezawa K, Pirazzoli V, Arcila ME, et al. HER2 amplification: a potential mechanism of acquired resistance to EGFR inhibition in EGFR-mutant lung cancers that lack the second-site EGFRT790M mutation. Cancer Discov 2012;2:922-33. [PubMed]
- Ohashi K, Maruvka YE, Michor F, et al. Epidermal growth factor receptor tyrosine kinase inhibitor-resistant disease. J Clin Oncol 2013;31:1070-80. [PubMed]
- Lev S, Yarden Y, Givol D. Dimerization and activation of the kit receptor by monovalent and bivalent binding of the stem cell factor. J Biol Chem 1992;267:15970-7. [PubMed]
- Linnekin D. Early signaling pathways activated by c-Kit in hematopoietic cells. Int J Biochem Cell Biol 1999;31:1053-74. [PubMed]
- Corless CL, Fletcher JA, Heinrich MC. Biology of gastrointestinal stromal tumors. J Clin Oncol 2004;22:3813-25. [PubMed]
- Yuzawa S, Opatowsky Y, Zhang Z, et al. Structural basis for activation of the receptor tyrosine kinase KIT by stem cell factor. Cell 2007;130:323-34. [PubMed]
- Gari M, Goodeve A, Wilson G, et al. c-kit proto-oncogene exon 8 in-frame deletion plus insertion mutations in acute myeloid leukaemia. Br J Haematol 1999;105:894-900. [PubMed]
- Tian Q, Frierson HF Jr, Krystal GW, et al. Activating c-kit gene mutations in human germ cell tumors. Am J Pathol 1999;154:1643-7. [PubMed]
- Curtin JA, Busam K, Pinkel D, et al. Somatic activation of KIT in distinct subtypes of melanoma. J Clin Oncol 2006;24:4340-6. [PubMed]
- Heinrich MC, Rubin BP, Longley BJ, et al. Biology and genetic aspects of gastrointestinal stromal tumors: KIT activation and cytogenetic alterations. Hum Pathol 2002;33:484-95. [PubMed]
- Goldman JM, Melo JV. Chronic myeloid leukemia--advances in biology and new approaches to treatment. N Engl J Med 2003;349:1451-64. [PubMed]
- Manley PW, Cowan-Jacob SW, Buchdunger E, et al. Imatinib: a selective tyrosine kinase inhibitor. Eur J Cancer 2002;38:S19-27. [PubMed]
- Hantschel O, Rix U, Superti-Furga G. Target spectrum of the BCR-ABL inhibitors imatinib, nilotinib and dasatinib. Leuk Lymphoma 2008;49:615-9. [PubMed]
- Tuveson DA, Willis NA, Jacks T, et al. STI571 inactivation of the gastrointestinal stromal tumor c-KIT oncoprotein: biological and clinical implications. Oncogene 2001;20:5054-8. [PubMed]
- Blanke CD, Rankin C, Demetri GD, et al. Phase III randomized, intergroup trial assessing imatinib mesylate at two dose levels in patients with unresectable or metastatic gastrointestinal stromal tumors expressing the kit receptor tyrosine kinase: S0033. J Clin Oncol 2008;26:626-32. [PubMed]
- Verweij J, Casali PG, Zalcberg J, et al. Progression-free survival in gastrointestinal stromal tumours with high-dose imatinib: randomised trial. Lancet 2004;364:1127-34. [PubMed]
- Dematteo RP, Gold JS, Saran L, et al. Tumor mitotic rate, size, and location independently predict recurrence after resection of primary gastrointestinal stromal tumor (GIST). Cancer 2008;112:608-15. [PubMed]
- Antonescu CR, Besmer P, Guo T, et al. Acquired resistance to imatinib in gastrointestinal stromal tumor occurs through secondary gene mutation. Clin Cancer Res 2005;11:4182-90. [PubMed]
- Heinrich MC, Corless CL, Demetri GD, et al. Kinase mutations and imatinib response in patients with metastatic gastrointestinal stromal tumor. J Clin Oncol 2003;21:4342-9. [PubMed]
- Corless CL, Barnett CM, Heinrich MC. Gastrointestinal stromal tumours: origin and molecular oncology. Nat Rev Cancer 2011;11:865-78. [PubMed]
- Nishida T, Kanda T, Nishitani A, et al. Secondary mutations in the kinase domain of the KIT gene are predominant in imatinib-resistant gastrointestinal stromal tumor. Cancer Sci 2008;99:799-804. [PubMed]
- Heinrich MC, Corless CL, Blanke CD, et al. Molecular correlates of imatinib resistance in gastrointestinal stromal tumors. J Clin Oncol 2006;24:4764-74. [PubMed]
- Antonescu CR. The GIST paradigm: lessons for other kinase-driven cancers. J Pathol 2011;223:251-61. [PubMed]
- Edkins S, O'Meara S, Parker A, et al. Recurrent KRAS codon 146 mutations in human colorectal cancer. Cancer Biol Ther 2006;5:928-32. [PubMed]
- Janakiraman M, Vakiani E, Zeng Z, et al. Genomic and biological characterization of exon 4 KRAS mutations in human cancer. Cancer Res 2010;70:5901-11. [PubMed]
- Douillard JY, Oliner KS, Siena S, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med 2013;369:1023-34. [PubMed]
- Allegra CJ, Jessup JM, Somerfield MR, et al. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol 2009;27:2091-6. [PubMed]
- Feig LA, Cooper GM. Relationship among guanine nucleotide exchange, GTP hydrolysis, and transforming potential of mutated ras proteins. Mol Cell Biol 1988;8:2472-8. [PubMed]
- Riely GJ, Kris MG, Rosenbaum D, et al. Frequency and distinctive spectrum of KRAS mutations in never smokers with lung adenocarcinoma. Clin Cancer Res 2008;14:5731-4. [PubMed]
- Marks JL, Broderick S, Zhou Q, et al. Prognostic and therapeutic implications of EGFR and KRAS mutations in resected lung adenocarcinoma. J Thorac Oncol 2008;3:111-6. [PubMed]
- Linardou H, Dahabreh IJ, Kanaloupiti D, et al. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. Lancet Oncol 2008;9:962-72. [PubMed]
- Jakob JA, Bassett RL Jr, Ng CS, et al. NRAS mutation status is an independent prognostic factor in metastatic melanoma. Cancer 2012;118:4014-23. [PubMed]
- Akslen LA, Puntervoll H, Bachmann IM, et al. Mutation analysis of the EGFR-NRAS-BRAF pathway in melanomas from black Africans and other subgroups of cutaneous melanoma. Melanoma Res 2008;18:29-35. [PubMed]
- Si L, Kong Y, Xu X, et al. Prevalence of BRAF V600E mutation in Chinese melanoma patients: large scale analysis of BRAF and NRAS mutations in a 432-case cohort. Eur J Cancer 2012;48:94-100. [PubMed]
- Ellerhorst JA, Greene VR, Ekmekcioglu S, et al. Clinical correlates of NRAS and BRAF mutations in primary human melanoma. Clin Cancer Res 2011;17:229-35. [PubMed]
- Lee JH, Choi JW, Kim YS. Frequencies of BRAF and NRAS mutations are different in histological types and sites of origin of cutaneous melanoma: a meta-analysis. Br J Dermatol 2011;164:776-84. [PubMed]
- Sosman JA, Kim KB, Schuchter L, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med 2012;366:707-14. [PubMed]
- Ascierto PA, Schadendorf D, Berking C, et al. MEK162 for patients with advanced melanoma harbouring NRAS or Val600 BRAF mutations: a non-randomised, open-label phase 2 study. Lancet Oncol 2013;14:249-56. [PubMed]
- Menzies AM, Long GV. New combinations and immunotherapies for melanoma: latest evidence and clinical utility. Ther Adv Med Oncol 2013;5:278-85. [PubMed]
- Menzies AM, Long GV. Systemic treatment for BRAF-mutant melanoma: where do we go next? Lancet Oncol 2014;15:e371-81. [PubMed]
- Ohashi K, Sequist LV, Arcila ME, et al. Characteristics of lung cancers harboring NRAS mutations. Clin Cancer Res 2013;19:2584-91. [PubMed]
- Xie P, Streu C, Qin J, et al. The crystal structure of BRAF in complex with an organoruthenium inhibitor reveals a mechanism for inhibition of an active form of BRAF kinase. Biochemistry 2009;48:5187-98. [PubMed]
- Wan PT, Garnett MJ, Roe SM, et al. Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell 2004;116:855-67. [PubMed]
- Thevakumaran N, Lavoie H, Critton DA, et al. Crystal structure of a BRAF kinase domain monomer explains basis for allosteric regulation. Nat Struct Mol Biol 2015;22:37-43. [PubMed]
- Davies H, Bignell GR, Cox C, et al. Mutations of the BRAF gene in human cancer. Nature 2002;417:949-54. [PubMed]
- Kimura ET, Nikiforova MN, Zhu Z, et al. High prevalence of BRAF mutations in thyroid cancer: genetic evidence for constitutive activation of the RET/PTC-RAS-BRAF signaling pathway in papillary thyroid carcinoma. Cancer Res 2003;63:1454-7. [PubMed]
- Tol J, Nagtegaal ID, Punt CJ. BRAF mutation in metastatic colorectal cancer. N Engl J Med 2009;361:98-9. [PubMed]
- Liu Y, Gray NS. Rational design of inhibitors that bind to inactive kinase conformations. Nat Chem Biol 2006;2:358-64. [PubMed]
- Chapman PB, Hauschild A, Robert C, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med 2011;364:2507-16. [PubMed]
- Dahlman KB, Xia J, Hutchinson K, et al. BRAF(L597) mutations in melanoma are associated with sensitivity to MEK inhibitors. Cancer Discov 2012;2:791-7. [PubMed]
- Poulikakos PI, Persaud Y, Janakiraman M, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 2011;480:387-90. [PubMed]
- Basile KJ, Abel EV, Dadpey N, et al. In vivo MAPK reporting reveals the heterogeneity in tumoral selection of resistance to RAF inhibitors. Cancer Res 2013;73:7101-10. [PubMed]
- Mao M, Tian F, Mariadason JM, et al. Resistance to BRAF inhibition in BRAF-mutant colon cancer can be overcome with PI3K inhibition or demethylating agents. Clin Cancer Res 2013;19:657-67. [PubMed]
- Hatzivassiliou G, Song K, Yen I, et al. RAF inhibitors prime wild-type RAF to activate the MAPK pathway and enhance growth. Nature 2010;464:431-5. [PubMed]
- Flaherty KT, Robert C, Hersey P, et al. Improved survival with MEK inhibition in BRAF-mutated melanoma. N Engl J Med 2012;367:107-14. [PubMed]
- Carvajal RD, Antonescu CR, Wolchok JD, et al. KIT as a therapeutic target in metastatic melanoma. JAMA 2011;305:2327-34. [PubMed]
- Minor DR, Kashani-Sabet M, Garrido M, et al. Sunitinib therapy for melanoma patients with KIT mutations. Clin Cancer Res 2012;18:1457-63. [PubMed]
- Daniels M, Lurkin I, Pauli R, et al. Spectrum of KIT/PDGFRA/BRAF mutations and Phosphatidylinositol-3-Kinase pathway gene alterations in gastrointestinal stromal tumors (GIST). Cancer Lett 2011;312:43-54. [PubMed]
- Zheng S, Huang KE, Pan YL, et al. KIT and BRAF heterogeneous mutations in gastrointestinal stromal tumors after secondary imatinib resistance. Gastric Cancer 2014. [Epub ahead of print]. [PubMed]
- Di Nicolantonio F, Martini M, Molinari F, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol 2008;26:5705-12. [PubMed]
- Prahallad A, Sun C, Huang S, et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 2012;483:100-3. [PubMed]
- Ohashi K, Sequist LV, Arcila ME, et al. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci U S A 2012;109:E2127-33. [PubMed]
- Corcoran RB, Ebi H, Turke AB, et al. EGFR-mediated re-activation of MAPK signaling contributes to insensitivity of BRAF mutant colorectal cancers to RAF inhibition with vemurafenib. Cancer Discov 2012;2:227-35. [PubMed]
- Andrews MC, Behren A, Chionh F, et al. BRAF inhibitor-driven tumor proliferation in a KRAS-mutated colon carcinoma is not overcome by MEK1/2 inhibition. J Clin Oncol 2013;31:e448-51. [PubMed]
- HaDuong JH. Sarcomas. Pediatr Clin North Am 2015;62:179-200. [PubMed]
- Mertens F, Tayebwa J. Evolving techniques for gene fusion detection in soft tissue tumours. Histopathology 2014;64:151-62. [PubMed]
- Li S, Qi X, Huang Y, et al. Ceritinib (LDK378): a potent alternative to crizotinib for ALK-rearranged non-small-cell lung cancer. Clin Lung Cancer 2015;16:86-91. [PubMed]


