Down syndrome and leukemia: insights into leukemogenesis and translational targets
Introduction
Children with Down syndrome (DS) have a significantly higher risk of developing leukemia in childhood as compared to children without DS (1), although curiously they have a lower risk of developing solid tumours (2). DS is defined by constitutional trisomy 21, which is the most common cytogenetic abnormality seen in live births, at a rate of 1/700 to 1/1,000 newborns (3,4).
The risk of developing acute megakaryoblastic leukemia (AMKL), which is a relatively rare subtype of acute myeloid leukemia (AML), is increased 500-fold in children with DS as compared to the general non-DS population; and risk of acute lymphoblastic leukemia (ALL) is 20-fold greater in children with DS (1).
Typically in childhood, ALL is significantly more common than AML. However in DS, the ratio of ALL to AML is 1.7 for children under 15 years of age. For the general population of non-DS children, the equivalent ratio is 6.5 (2).
In this review, we will focus on recent studies that have improved our understanding of leukemogenesis in DS, particularly myeloid leukemia of Down syndrome (ML-DS). We will also highlight important developments likely to translate into improved clinical treatment of ALL associated with DS (DS-ALL). Specifically we aim to identify potential impacts of new research on how we manage children with DS, pre-leukemia and leukemia.
Myeloid Leukemia of Down syndrome (ML-DS)
ML-DS includes DS-AMKL and myelodysplastic syndrome (MDS) (5). MDS often precedes DS-AMKL. In this paper, we will refer to both entities (DS-AMKL and DS-related MDS) as “ML-DS”.
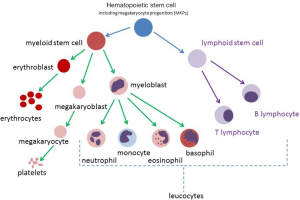
DS-AMKL, a sub-type of AML, is characterised by an abnormal monomorphic population of circulating megakaryoblasts. AML refers to a broader category of blood cancers that are derived from myeloid precursors. Megakaryoblasts are derived from myeloid precursors (Figure 1). Normal megakaryoblasts will differentiate into megakaryocytes (MKs) (platelet-producing cells). Abnormal megakaryoblasts will overwhelm normal bone marrow production and reduce production of the other cell lines, such as leucocytes (including neutrophils) and erythrocytes.
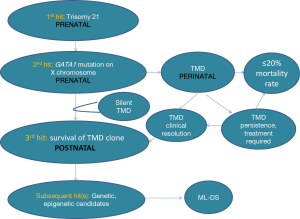
ML-DS is characterised by transforming events that occur in the fetal and newborn period (6). There are two cytogenetic and genetic changes or “hits” that occur prenatally, that give rise to a pre-leukemic state, called transient myeloproliferative disorder (TMD). TMD is also referred to as transient abnormal myelopoiesis (TAM) or transient leukemia (TL). The first hit is the presence of trisomy 21, which leads to increased proliferation of megakaryocyte progenitors (MKPs) in the fetal liver. The subsequent transforming event is a mutation in GATA binding protein 1 (GATA1) which gives rise to TMD. The third and subsequent hits are as yet unknown (6).
Transient myeloproliferative disorder (TMD)
TMD can occur in DS; in children without DS but with acquired somatic trisomy 21 mutations; and also mosaic trisomy 21 patients (7-10). There may be other leukemogenic factors, as yet unknown; as hypothesised in a recent case report of a newborn with clinical TMD, who did not have trisomy 21 or a GATA1 gene mutation in the leukemic blasts (7).
TMD occurs in at least 5-10% of newborns with DS, and is evidenced by presence of circulating megakaryoblasts, that are indistinguishable from blasts seen in ML-DS. Typical features of TMD include circulating peripheral megakaryoblasts with the immunophenotype CD33/38/117/34/7/56/36/71/42b (8), thrombocytopenia, variable presence of leucocytosis and anemia, hepatomegaly, splenomegaly, serous effusions and sometimes skin rash (9). Flow cytometry performed on peripheral blood can help characterise the lineage-specific markers present in the blast population. TMD can present in utero with severe hydrops fetalis.
Three prospective series found a median TMD diagnosis at 3-7 days postnatally and almost all cases had presented by age 2 months (8). Clinical features reflect transient abnormal megakaryopoiesis, which is thought to primarily originate in the liver (8,9). This is evidenced by the role of the fetal liver in hematopoiesis, and the generally lower presence of blasts in the bone marrow as compared to the peripheral blood (9).
Historically, TMD diagnoses have been made based on a full-blood count (FBC) being performed in a symptomatic DS newborn and the subsequent finding of megakaryoblasts in the peripheral blood. Currently there is no routine use of molecular testing of samples from TMD patients for GATA1 mutations; and not all newborns with DS will have a FBC in the newborn period. This means that there is likely to be a population of newborns with DS who have subclinical TMD.
In the majority of cases, TMD will resolve spontaneously, usually within the first 3 months of life. Treatment may be required in the event of life-threatening symptoms from TMD (such as respiratory or liver impairment). In total, 85-90% of newborns with TMD will demonstrate resolution of TMD, either spontaneously or due to therapeutic intervention (11-13). Therapy for TMD, although not commonly given, consists of low dose cytarabine. There is a mortality rate of up to 20% for patients with TMD (8). Event-free survival rates are approximately 60%, due to death and leukemic relapse (11,13). Early death and poor event-free survival are both predicted by hyperleukocytosis, severe liver dysfunction, prematurity and failure of spontaneous TMD remission (11). Coagulopathy and renal failure are additional poor prognosticators (11).
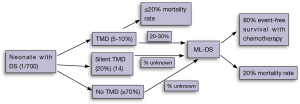
In 20-30% of cases, newborns with a history of TMD will go on to develop ML-DS, within the first 2-4 years of life (8) (Figure 2). Overall, between 0.5-2% of children with DS will develop ML-DS (2,15). The lack of universal testing of newborn children with DS for TMD and GATA1 mutations means that some newborns with DS and a significant risk of ML-DS in future, do not come to immediate clinical attention in the newborn period.
Unanswered questions in the field are: (I) What is the true incidence of subclinical (silent) TMD? (II) What is the clinical pattern of detectable GATA1 mutations over the period between TMD and ML-DS or resolution? (III) How does the GATA1 loss-of-function mutation contribute to the pre-leukemia, TMD, or the frank leukemia, ML-DS? (IV) Can children with DS who will progress to ML-DS be identified before frank leukemia develops; (V) Can progression from TMD to ML-DS be prevented?
The true incidence of silent TMD
Until recently, the concept of silent TMD was not well understood, however novel research findings have generated new hypotheses (14). Earlier studies found that the incidence of TMD in newborns with DS was between 3.8-6% (15,16). One of these studies used FBC results from all DS children within the first week of life (16), whilst another retrospectively analysed DS neonatal blood spots for GATA1 mutations (15). Only those with a GATA1 mutation at birth progressed to ML-DS (15). Many of these earlier studies were limited by sensitivity of PCR techniques that were available at the time (15).
In a recent population-based study of 200 neonates with DS, Roberts et al. have proposed that up to 30% of newborns with DS will have a detectable GATA1 gene mutation, if both conventional and next-generation sequencing approaches are used (14). Next-generation sequencing of exon 2 of GATA1 was performed on 104 patients. Newborns with a peripheral blast count of >10% and a detectable GATA1 gene mutation by conventional (Sanger Sequencing/Denaturing High Performance Liquid Chromatography) techniques have been labelled as “TAM” or “TMD”. Those newborns with a GATA1 gene mutation detectable only by next-generation sequencing are termed “silent TAM” or “silent TMD”. In this study all patients with silent TMD had a peripheral blast count of ≤10%. Newborns with peripheral blasts at birth, but no detectable GATA1 mutation were categorised as “no TMD” (Figure 2).
Therefore, the true incidence of TMD (including asymptomatic or “silent TMD” cases) may lie between 10-31%, depending on definitions used (9,11-14). The clinical implications for those children with “silent TMD” is uncertain, however these children do have an increased risk of subsequent ML-DS (14). In the recent prospective UK cohort, 11% of newborns with GATA1 mutations (including those with TMD and silent TMD) later developed ML-DS (14).
Currently however, GATA1 gene mutation testing is not standard of care in newborns with DS. International guidelines recommend at least a FBC to be performed at least once before 1 month of age (17). Many institutions will perform a FBC at birth. Regular blood tests are recommended to monitor for systemic manifestations of DS, including yearly FBC for macrocytic anaemia and more frequent monitoring of thyroid function (17).
Children who have documented TMD require monitoring for resolution of the disease, then more frequent monitoring, due to their increased risk of ML-DS. In our own clinical practice, we perform a FBC and clinical review every 3 months for these children, up until age 4 years. This is consistent with international practice (8). After 4-5 years of age, the incidence of leukemia is significantly less (2).
Treatment and outcome for ML-DS
Children treated for ML-DS have a significantly higher disease-free survival (DFS) compared to other children treated for AML (DFS 88-89% compared to 42%, P<0.001) (18,19). Arguably this could be due to AML subtype but DS-AMKL also requires less intense therapy to achieve cure as compared to non-DS AMKL, indicating that children with ML-DS are more responsive to chemotherapy (18,19). A possible explanation for the chemosensitivity of ML-DS is an alteration in cytarabine drug metabolism (20,21) due to reduced cytidine deaminase gene expression in ML-DS (21) or increased expression of cystathionine β-synthase, which is encoded by chromosome 21 (20). Cytarabine is a key drug in successful therapy against ML-DS and AML.
Children with DS who develop AMKL beyond the age of 4 years old are thought to represent a different cohort of patients. Age >4 years old is a poor prognosticator, conferring a 5-year EFS of 33%, compared to 81% for DS children with myeloid leukemia aged <4 years old (19). Children who are older than 4 years of age, without GATA1 mutations, are more likely to have similar cytogenetic aberrations to sporadic (non-DS) AML (22). These children are more likely to require more intensive therapy, as compared to children with ML-DS (22). In addition, DS patients with TMD have also been described who progress to ALL, although this is very rare; and occurs much less often than ML-DS (12,13).
Multi-step process of leukemogenesis in ML-DS: role of GATA1 loss-of-function mutation in the pre-leukemia, TMD, or the frank leukemia, ML-DS
The effect of trisomy 21 and GATA1 mutation in promoting abnormal megakaryopoiesis has been recently clarified. The acquisition of trisomy 21 alone is the first hit, as trisomy 21 without GATA1 mutation leads to altered myeloid progenitor self-renewal, altered lineage development (23,24) and increased clonogenicity of MKPs in human fetal livers (25).
Somatic GATA1 mutation is identified as the “second hit”, and it is thought to block MK differentiation (26). The GATA1 gene mutation has been identified in almost all cases of TMD and ML-DS. GATA1 somatic mutation was first identified from a small series of ML-DS samples (26). GATA1 is located on the X chromosome; and encodes a zinc finger-containing protein that is essential for normal erythropoiesis and megakaryopoiesis (26,27). GATA1 gene mutation leads to sole production of a truncated GATA1 protein, called GATA1s. GATA1s lacks an amino-transactivation domain but retains both DNA-binding zinc fingers (26). The majority of mutations have been described in exon 2, with a minority in exon 3 or at the intronic boundary of exon 1 and 2. In 75% of cases these are insertions, deletions or duplications. Point mutations are described in 21% of cases (4). The presence of GATA1s is thought to impair GATA1-mediated regulation of other transcription factors, including GATA2, MYB, MYC and IKAROS family zinc finger 1 (IKZF1) in fetal MKs (28).
GATA1 mutations, in the absence of trisomy 21, have not been associated with leukemia. Instead, specific hematopoietic alterations due to GATA1 mutation alone include cytopenias (26), Diamond-Blackfan anaemia (29) and trilineage bone marrow dysplasia in germline GATA1 mutation (30). Therefore GATA1 mutation represents the second hit, in the presence of trisomy 21.
Analysis of paired samples from the same patient has found the identical GATA1 gene mutation in both the pre-leukemia (TMD) and leukemia (ML-DS) (11,27,31,32). It is highly likely that additional transforming events, or “hits”, are involved in this leukemogenic process (6,33,34). Current knowledge regarding the ability to detect and quantify GATA1s or GATA1 mutations from a clinical TMD episode through to either TMD resolution or evolution to ML-DS is limited. These additional ‘hits” may be genetic and/or possibly epigenetic changes (Figure 3). In one study, 44% of ML-DS samples demonstrated additional genetic mutations, aside from trisomy 21 and GATA1 mutation (33). Additional “hits” may contribute to the survival of pre-leukemic cells in the postnatal environment and play a role in subsequent risk of leukemic transformation within the bone marrow compartment.
Modelling of DS and ML-DS
Several methods have been used to study clonal evolution and additional “hits”. The most relevant studies to date have analysed primary patient samples including paired patient samples as well as mouse models of leukemogenesis; DS fetal tissue and induced pluripotent cells derived from fetal tissue. These are described in the next section.
Paired patient samples may provide an indication of clonal evolution, although clonal evolution is not always seen (11). Examples of clonal evolution, comparing TMD to ML-DS samples from the same patient, include additional genetic alterations such as trisomy 8 (27) and AML-associated changes such as der (3q), trisomy 19 and trisomy 11 (11). This indicates that there is abnormal persistence of a blast population, that subsequently gives rise to the definitive ML-DS state. This also implies that residual TMD clones could be detected by minimal residual disease (MRD) techniques such as polymerase chain reaction (PCR), flow cytometry, or next-generation sequencing. Use of semi-quantitative PCR has been recently reported for 2 patients (35). One patient had GATA1 mutation analyses performed retrospectively after detection of the GATA1 mutation in the ML-DS sample. This demonstrated presence of GATA1 mutation from age 3 months (silent TMD) and persistence until ML-DS diagnosis. The other patient had TMD and reduction in the size of the GATA1 mutated clone over subsequent months.
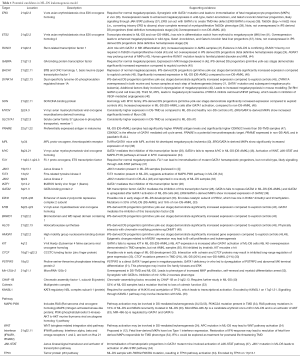
Several studies have attempted to define candidate genes, including genes on human chromosome 21, as potential effectors of leukemogenesis in DS. A summary of potential genes and drivers in DS-mediated leukemogenesis is listed in Table 1. Differing pathways for leukemogenesis may exist in DS-AMKL and non-DS-AMKL, as oncogene expression varies between the two entities (40,46).
Key candidate genes on chromosome 21 include ERG, ETS2, RUNX1, GABPA [reviewed in (4)], BACH1 (8) and DYRK1A (40).
Members of the ETS gene family, ERG, ETS2 and FLI1 have been shown to contribute to dysregulation of megakaryopoiesis in fetal liver progenitors in GATA1 mutant mice (37). ERG and ETS2 have a proliferative effect on MKs, independent of GATA1 (37). ERG and FLI-1 both lead to immortalisation of hematopoietic progenitor cells in GATA1 mutant mice, probably through JAK/STAT pathway activation (37).
Confirmatory studies implicate ERG in dysregulated megakaryopoiesis in GATA1 mutant mice models (37), in immortalisation of hematopoietic progenitors (36,37) and as contributor to leukemogenesis in adult bone marrow cells (55). Perhaps the most striking observation is that ERG can cooperate with GATA1s to create a TMD-like defect in vivo and potentially lead to myeloid leukemia (38). ERG alone led to increased MKPs in fetal liver (38). Earlier models, with large triplicated regions of orthologues found on human chromosome 21 (Hsa21), failed to demonstrate myeloproliferation despite triplicated copies of ERG (56). Modulation of the Ts65Dn mouse, which converted the mice from ERG trisomy to ERG disomy [Ts65Dn(Erg+/+/mld2)], led to a complete reversal of the myeloproliferative phenotype observed in Ts65Dn mice (39). Studies using human cell lines have demonstrated that ERG (an ETS transcription factor) is found in hematopoietic stem cells (HSCs), megakaryocytic cell lines and primary leukemia cells in DS (41).
ERG can alter HSC and megakaryocytic development by switching differentiation from erythroid to megakaryocytic phenotypes, through activation of gp1b and gpIIb promoters in vivo; and, together with ETS2, by binding the hematopoietic enhancer SCL/TALI in vivo (41).
In addition, ERG-expanded adult bone marrow T-cells with an added NOTCH1 gene mutation, developed ALL (55). However, ERG-expressing pro-B cells in culture but could not induce B-cell leukemia (55). ERG expression is a poor prognosticator in adult T-ALL and cytogenetically-normal AML, although its role in childhood leukemias is uncertain (55). A crucial role in maintenance of leukemia in adult HSCs is shown by shRNA knockdown of ERG, which resulted in reduced cell growth in erythroid, myeloid, T and B cells (55).
RUNX1 and DYRK1A genes lie in the DS Critical Region (DSCR), a region at 21q22 that is thought to be responsible for DS phenotypic features.
RUNX1 is an aetiologic factor in AML (8) although there is conflicting evidence regarding RUNX1 in ML-DS (Table 1). Human germline RUNX1 mutation leads to familial platelet disorder and AML (44). Results from Tc1, Ts65Dn and Ts1Cje mouse studies would indicate that RUNX1 does not cause abnormal hematopoiesis (57), myeloproliferation (43) or MKP expansion (56). However, there may be a role for RUNX-1 in potentiation of megakaryopoiesis, as survival of the MKPs was longer in trisomic RUNX-1 mice (43). GATA1 interacts with RUNX1 to facilitate normal megakaryopoiesis in cell lines (42). RUNX1 accounted for some of the increased gene expression observed in DS-hyperproliferation in vitro, including a significant increase in c-Kit and Tie-2 expression (58). The mechanism by which altered GATA1s/ RUNX1 interaction may lead to ML-DS is unclear (42).
DYRK1A is overexpressed in DS-TMD and ML-DS (40). DYRK1A expression leads to increased megakaryopoiesis and inhibition of the calcineurin/NFAT (nuclear factor of activated T-cells) pathway (40). Dysregulation of the calcineurin/NFAT pathway is thought to account for increased leukemic incidence and reduced solid tumour incidence in DS (40).
MicroRNAs encoded by chromosome 21 may also play a role in leukemogenesis (8). miR-125b-2 is overexpressed in ML-DS (DS-AMKL) and DS-TMD, is found to increase proliferation and self-renewal of MKPs; and causes a myeloid differentiation arrest (52). miR-125b-2 and GATA1s synergistically increased proliferation of relevant hematopoietic precursors, and inhibition of miR-125b-2 caused impaired growth in DS-AMKL/DS-TMD (52). MicroRNA-486-5p, on chromosome 8, has recently been shown to be regulated by both GATA1 and GATA1s; and to act as an erythroid onco-miR in ML-DS (53).
Epigenetic targets on chromosome 21 include BRWD1, HLCS, and HMGN1 (45,49) (Table 1).
Other candidate drivers not encoded by chromosome 21 include JAK3 (4), MYCN (47), MYC (34), PRAME (47) and additional epigenetic modifiers such as EZH2 (Table 1). GATA1-target genes identified through non-DS models may also reveal candidates, such as PSTPIP2 (51).
Full table
Additional models to study ML-DS
Development of a DS TMD/ML-DS model remains a challenge. One group was successful in creating AMKL mouse xenografts, however a mouse model of DS-TMD evolving to ML-DS has not been described (59). Promising models to date include one that mimics the function of GATA1s (28) and a model of ERG/GATA1s mice that developed a TMD-like expression profile and MKP proliferation defect (38).The ERG/GATA1s mice were reported to develop myeloid leukemia at 3 months (38). A recent xenograft incorporating patient-derived TMD samples demonstrated emergence of genomic features of ML-DS after serial transplantation. These genomic changes were present in small subclones in the original TMD samples (60).
Well studied murine models of DS include Tc1, Ts65Dn, Ts1Cje (43,56,57) and GATA1 knock-in mice (28). Ts1Rhr mice have also been used to study DS phenotypes, including ML-DS (40) and DS-ALL (61). These models differ in the number of relevant triplicated genes present; and this in part depends on which mouse chromosomes (and thus orthologues) are present (62). The Ts65Dn and Ts1Cje mouse models use mouse chromosome 16 (Mmu16), on which the majority of Hsa21 genes are located (56). Mouse chromosome 10 and 17 contain the remainder of Hsa21 orthologues (57).The most widely used model, Ts65Dn model, is trisomic for the distal end of chromosome 16q, and this is fused to a genetically-poor region of 17p (43). Tc1 is trisomic for 269 genes orthologues on Hsa21 whilst Ts65Dn includes 104 gene orthologues, 94 of which are from the DSCR (Hsa21q22) (43,57). The Ts1Cje mouse has 97 orthologues and Ts1Rhr has 31 orthologues, analogous to part of the DSCR (56,61). Tc1/GATA1Δe2 double mutant mice did not develop TMD or leukemia, despite this model offering the largest number of Hsa21 orthologues to study hematopoiesis (57). Only the Ts65Dn model developed a myeloproliferative phenotype, albeit progressive and occurring at 15 months of age (43). Therefore, although useful, this model does not replicate human ML-DS.
The role of GATA1s in myeloproliferation has been studied in mouse models. The GATA1 knock-in model (GATA1ΔN) confirmed a megakaryocytic proliferative phenotype in yolk sac and fetal liver hematopoiesis, that was sensitive to the effects of GATA1s (28). The GATA1Δe2 model closely replicates the GATA1s truncated protein that is produced by N-terminal GATA1 gene disruption. MK proliferation occurred, without an accompanying differentiation block, possibly through loss of inhibition of growth regulatory genes (28). N-terminal disruption of GATA1 can be compensated for by signalling through the C-terminal transactivation domain (C-TAD) of GATA1, and C-TAD has a role in embryonic hematopoiesis and megakaryopoietic proliferation (63).
Due to limitations of using mouse models, including the lack of a TMD phenotype with spontaneous resolution, other hematopoietic systems have been studied. Recent work has focused on in vitro differentiation of isogenic human pluripotent cells. In vitro studies of fetal DS (24,45) report varying gene expression results, according to the stage of fetal hematopoiesis. Characteristic MKP proliferation was detected in fetal DS hematopoiesis (24), specifically definitive fetal hematopoiesis only (23). In addition, Roy et al. demonstrated impaired B-lymphoid differentiation in DS fetal liver tissue (23). BACH1, GABPA, SON, DYRK1A were significantly up-regulated in DS progenitor cells in early fetal hematopoiesis, compared to euploid human cell lines (45). No alteration of ETS, ERG, RUNX-1 or IGF signalling genes were found in definitive fetal hematopoiesis in human DS samples (24). In vitro studies are limited by the small magnitude of gene expression changes that are detected, therefore stage-specific changes would ideally be confirmed by additional disease models.
Can children with DS who will progress to ML-DS be identified before frank leukemia develops?
The most likely candidate markers for DS patients at risk of progressing to ML-DS are the GATA1s protein itself or the various mutations which cause this protein to be prematurely truncated. The GATA1s level in peripheral blood or bone marrow at the diagnosis of TMD and its pattern of change over time, which predicts the subsequent risk of ML-DS, is unknown. GATA1 mutations are restricted to blast cells, and therefore clear after resolution of TMD or ML-DS (15,27,64,65). Therefore, the true prevalence of GATA1 mutations in DS, and the timing of re-emergence of the GATA1 mutant clone remain unanswered questions. There is debate regarding whether the type of GATA1 mutation predicts progression to ML-DS. The largest study to date of TMD (134 samples) and ML-DS (103 samples) did not show any correlation between type of GATA1 mutation and progression to ML-DS (32). However, another study found a surprising association between low GATA1s protein levels, associated with particular types of GATA1 mutations, and a significantly higher risk of ML-DS (66).
To definitively answer these questions, a prospective, longitudinal study of all DS patients is needed. With current next-generation sequencing techniques we may be able to further elucidate drivers and repressors of leukemogenesis, using matched TMD and ML-DS samples. To our knowledge, there are no published prospective longitudinal data that have evaluated the pattern of GATA1 gene mutations and resultant gene expression over time. In our own research study, the PreP21 (Predicting and Preventing Leukemia in Children with DS) study (Clinical trials reference number: ACTRN12613000861752), we aim to study longitudinal changes over time in patients with TMD. Prospective sampling will allow detection of subclinical TMD. GATA1 mutations and additional drivers for leukemogenesis may be assessable in a longitudinal manner, in a similar manner to MRD monitoring in ALL therapy. This may then become a reliable biomarker for risk of progression to ML-DS. We hope to identify factors that promote regression of TMD in DS patients, or subsequent progression to ML-DS. In addition, we may be able to identify molecular targets for therapeutic targeting to prevent DS-mediated leukemogenesis.
Can progression from TMD to ML-DS be prevented?
Three prospective studies (POG, BFM, COG) found an incidence of 19-23% for DS patients with TMD developing ML-DS at a later stage (9). This occurred at a median time of 1.2-1.5 years from TMD (9). COG trial A2971 prospectively enrolled DS-TMD and ML-DS patients, and found that 43% of patients with ML-DS had prior TMD (19). The trial was run before PCR studies on GATA1 mutations revealed its importance, thus no GATA1 analysis was performed.
Therapeutic intervention for patients with TMD has been undertaken in small studies. Low dose cytarabine was used for babies who were symptomatic from severe TMD, with dose ranges varying from 1.2-1.5 mg/kg/dose twice daily for 7 days (subcutaneous or intravenous) (12) or 0.5-1.5 mg/kg for 3-12 days (11). Higher doses (3.33 mg/kg/day for 5 days) used in another trial led to Grade 3-4 toxicity in 96% of patients, without a discernible survival improvement (13).
Klusmann et al. studied 146 newborns with TMD and found that cytarabine therapy improved outcomes for those with risk factors for early death (11). Cytarabine was administered to those DS patients with TMD and clinical compromise due to high white cell count, thrombocytopenia or liver dysfunction. Patients with TMD who were treated with cytarabine developed ML-DS at the same frequency as those with TMD who did not received cytarabine, although numbers of treated patients were small (n=28). Patients in this study who went on to develop ML-DS with a history of TMD (n=29) had a significantly higher EFS than those without a history of TMD who developed ML-DS during the same period of study (n=142) (EFS 91% compared to 70%, P=0.039) (11).
Taken together, prior evidence suggests that treatment of TMD does not prevent progression to ML-DS, based on three prospective studies [reviewed in (9)]. Previous studies were not always able to test for GATA1 mutations; and if GATA1 was analysed, there were limitations in sensitivity of GATA1 mutation detection techniques and adequate samples. There is one current study (EudraCT no. 2006-002962-20) which aims to assess feasibility of low-dose cytarabine therapy to prevent progression from TMD to ML-DS by eradication of GATA1s and use of MRD monitoring (62). Therefore further study is required, in particular incorporating routine, sensitive GATA1 mutation testing.
Novel therapies in ML-DS
Therapeutic targets for further study in ML-DS include c-Kit inhibition using imatinib (50), miR-125b-2 inhibition (52) and interferon therapy (54). Low dose interferon may restore inhibition of MKP proliferation (54) and may therefore be an effective treatment strategy for residual MKPs in TMD (6).
Recent pre-clinical work has elucidated other potential drug therapies, such as wee1 kinase inhibitor MK-1775 in ML-DS (67) and Aurora–A Kinase (AURKA) inhibitors [such as dimethyl fasudil and MLN8237(59)] in AMKL. MK-1775 enhanced cytarabine-induced cytotoxicity in vitro in ML-DS cell lines and in ex vivo primary patient samples (67). AURKA inhibitors lead to polyploidisation, mature cell-surface marker expression and apoptosis of malignant (non-DS) AMKL cells (59). The role for treatment of ML-DS with AURKA inhibitors has not yet been established.
Understanding of GATA1 biology in ML-DS can be applied to non-DS AML. For example, elegant pre-clinical studies demonstrated that high GATA1 expression correlated with increased Bcl-xl protein levels (68). Knock-down of GATA1 in megakaryocytic cell lines partly reduced Bcl-xl expression, resulting in increased apoptosis and increased chemosensitivity. There is a Bcl-2 inhibitor GX15-070 (obatoclax) currently in early phase clinical trials in leukemia (68). Sodium valproate, in the same study, downregulated GATA1 expression and led to enhanced cytarabine-induced apoptosis in vitro. Therefore Bcl-2 inhibitors and sodium valproate, a histone deacetylase inhibitor, are potential therapeutic agents in non-DS AML with high GATA1 expression (68).
ALL in DS: “DS-ALL”
This type of leukemia, derived from lymphoid precursors, is more common in the general (non-DS) population than AML. The most common types of ALL we see in paediatric practice are B-lymphoblastic leukemia (B-ALL), affecting B-lymphocytes; T-lymphoblastic leukemia (T-ALL, T-lymphocytes) and mixed phenotype acute leukemia (MPAL) (69).
We will discuss DS-ALL in the context of new translational targets that may be used for future treatment strategies. The treatment landscape in DS-ALL is very different to ML-DS.
Treatment and outcome for DS-ALL
In the non-DS population, the overall event-free survival (EFS) for ALL is 85% or greater (70). In contrast, overall survival (OS) for children with DS-ALL is closer to 70% compared to 89% for non-DS ALL (P<0.0001) (71). Increased chemosensitivity documented in ML-DS cells has not been observed in DS-ALL cells (8).
Genetic features that may have an adverse impact on overall survival from DS-ALL include JAK2 mutations (that are found in 20% of DS-ALL) (48) and aberrant expression of the type 1 cytokine receptor CRLF2 in 60% of children with DS-ALL (72). CRLF2-positive DS-ALL is likely to be classified as high-risk ALL if present in combination with IKAROS (IKZF1) gene deletion (71). Recently, RAS driver mutations (KRAS, NRAS) were identified in 1/3 cases of DS-ALL; occurring virtually exclusively of, and at a similar frequency to, JAK2 mutations (73). Potential good prognostic features include ETV6/RUNX1 fusion and high hyperdiploidy (71). Other factors that influence OS include increased rate of treatment-related toxicity and increased risk of infectious deaths compared to non-DS ALL (74,75). Treatment-related toxicity, for example due to methotrexate, may be explained by altered metabolic profiles in non-leukemic cells, caused by constitutional trisomy 21 [reviewed in (76)].
Potential reasons for the increased rate of infectious deaths in children with DS-ALL, which can occur during less intensive maintenance therapy, are firstly an immunodeficient state characterised by partial B-cell deficiency with resulting dysgammaglobulinemia and secondly, dysregulation of T-cell function due to inhibition of the calcineurin/NFAT pathway [reviewed in (74)]. Epigenetic studies indicate this may be due to reduced expression of proteins involved in cell signalling pathways, such as cytokine-cytokine receptor interaction pathways (61).
Two recent studies reported that children with DS-ALL still have a high risk of relapse, even after hematopoietic stem cell transplantation (HSCT) (71). Therefore the challenge is identifying those children with high-risk DS-ALL who require treatment intensification to prevent subsequent relapse; and correctly identifying those children with DS-ALL who have favourable molecular features that will permit treatment de-intensification and still achieve lasting remission (74,77). MRD response could be used to discriminate DS-ALL patients with a low-risk of relapse and to intensify treatment for DS-ALL with a poor MRD response (75). Successful use of MRD-risk directed therapy was described for a large cohort of children with BCR-ABL1-like ALL, which included some children with DS (78). The heterogeneity of DS-ALL, with respect to biological features and treatment response, remains a clinical challenge.
Drivers of leukemogenesis in DS-ALL
The relevance of recent identification of altered JAK-STAT and RAS signalling in DS-ALL has therapeutic potential. JAK-STAT inhibitors are already in clinical use for adults with myelofibrosis (77) and systematic trials in paediatric DS-ALL are awaited (74). JAK-STAT inhibitors, such as the dual JAK1/JAK2 inhibitor ruxolitinib, may in future be used as adjunct therapy for children with CRLF2 positive-ALL to induce remission (74) and provide a bridge to HSCT for high-risk paediatric DS-ALL. A recently developed KRAS inhibitor, deltarasin, may be of benefit to patients with DS-ALL and RAS-mutations (73); and this would need to be studied in early phase trials.
One potential translational research question is whether there are any pre-leukemic initiating events that occur in DS-ALL, similar to ML-DS. Of interest, is the known in utero transforming event of ETV6/RUNX1 (TEL-AML1) fusion that can be detected postnatally and leads to increased leukemogenic potential of the transformed B-cells (6,79). RUNX1, as previously discussed, is located on chromosome 21 and has a role in megakaryopoiesis. A recent study implicated mir-125-b2 as a potential independent driver in ETV6/RUNX1 non-DS ALL (80). However, in DS-ALL, there is a decreased prevalence of both favourable (e.g., ETV6/RUNX1) and unfavourable chromosomal aberrations (e.g., BCR-ABL) (74,81), suggesting that there may be a different driver of leukemogenesis in DS-ALL.
DS and leukemia: epigenetics and future directions
Epigenetics may also play a broad role in DS-ALL and ML-DS. Two recent studies used Ts1Rhr mouse models to analyse epigenetic changes, the first in ML-DS (82) and the second study in DS-ALL (61).
Malinge et al. found that trisomy 21 led to global hypomethylation; and that DS-TMD samples featured new, focal gains of DNA methylation. Hypomethylation of the DSCR in particular may lead to increased expression of trisomic genes that predispose to DS-mediated myeloid leukemogenesis (82). In contrast, the transcriptome and epigenome of DS-TMD samples compared to ML-DS samples were very similar (82).
In DS-ALL, a transcriptional profile was defined based on analysis of B-lymphocytes (61). The analyses revealed highly enriched clustering in pathways related to polycomb repressor 2 (PRC2) targets and sites of trimethylated Lys 27 of histone 3 (H3K27me3). H3K27me3 is the repressive epigenetic mark added by PRC2. DS-ALL demonstrates global reduction in H3K27me3, which in turn leads to an increased gene expression pattern that drives B-cell development. By using a histone demethylase inhibitor, GSK-J4, H3K27me3 expression was increased in Ts1Rhr B cells, and led to reversal of the B-cell leukemogenic phenotype (61). The study found a potential candidate HMGN1 in Ts1Rhr mice that resulted in global suppression of H3K27me3. Sole HMGN1 overexpression in shRNA modified- Ts1Rhr mouse models led to phenotypic changes seen in the normal Ts1Rhr mice, therefore providing further proof that HMGN1 is largely responsible for the B-cell leukemogenic changes. The authors provide a proof of principle that by reversing global suppression of H3K27me3, which is likely due to HMGN1, B-cell leukemogenesis may be blocked (61).
An elegant study of monozygotic twins, discordant for trisomy 21, also describes profound genome-wide changes associated with trisomy 21. The additional chromosome 21 is thought to promote changes in the transcriptome of trisomic cells, affecting protein-coding genes and lncRNAs (49). These transcriptional changes were organised in well-defined chromosomal domains, termed “gene expression dysregulation domains” (GEDDs). GEDDs were replicated in additional models, including induced pluripotent stem cells derived from fibroblasts from the discordant twins; and the Ts65Dn mouse model (49). GEDDs may be the result of Hsa21-based genes that modify the chromatin environment of the nuclear compartments in trisomic cells. Candidate genes on chromosome 21 that may contribute to epigenetic changes include holocarboxylase synthetase (HLCS), and proteins HMGN1, DYRK1α, RUNX1, BRWD1 (49). This hypothesis requires further study, including whether the transcriptional changes are due to chromosome 21-based candidates; or as a general result of any human trisomy. These findings could then be applied to both DS-ALL and ML-DS.
Lastly, targeting pre-leukemic cells and eradicating leukemic stem cells that account for relapse will be a major challenge. Potentially, further study could assess whether an early HSC precursor is mutated, prior to lymphoid/myeloid pathway commitment (58). This is plausible, due to HSC expansion induced by trisomy 21 alone. Genetic alterations in a pluripotent HSC could explain the increased incidence of both myeloid and lymphoid malignancies in children with DS; and also explain rare cases of non-contemporaneous AML and ALL occurring in the same individual (74).
Conclusions
Therefore, the ultimate aim is to identify novel therapeutic targets that may improve outcome for all children with DS, pre-leukemia and leukemia. Insights into TMD/ML-DS may help us understand how early fetal hematopoietic development promotes leukemogenesis in disparate patient populations and may help us understand the aetiology of DS-ALL. Prospective GATA1 analysis may provide a platform for identification and intervention, to prevent ML-DS and improve quality of life in children with DS. Robust DS-TMD/ML-DS xenograft models will help to define the overall role of GATA1s and to understand the elusive third and subsequent “hits”. Xenograft models may also promote further understanding of chemosensitivity and, inversely, resistance of DS-leukemia cells. Translational models of ML-DS and DS-ALL will permit dynamic analysis of Hsa21 genes and provide a platform for development of targeted agents for high-risk leukemia. Moreover, this knowledge may collectively provide insights that may be applied to non-DS leukemia and possibly all embryonal cancer.
Acknowledgements
Funding: GM Marshall is supported by grants from the Australian National Health and Medical Research Council (NHMRC); the Cancer Institute New South Wales, Australia; the Cancer Council New South Wales, Australia; and the Steven Walter Children’s Cancer Foundation. R Sutton is supported by grants from the NHMRC. MK Mateos receives support through a Kids Cancer Project Research Entry Scholarship administered via the Royal Australasian College of Physicians. We also gratefully acknowledge the Kids with Cancer Foundation for financial support of the Kids Cancer Centre; and the Sydney Children’s Hospital Foundation. Sydney Children’s Hospital, Randwick, New South Wales, Australia and the Children’s Cancer Institute Australia for Medical Research, Randwick, New South Wales, Australia, are affiliated with the University of New South Wales, Sydney, Australia.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Hitzler JK, Zipursky A. Origins of leukaemia in children with Down syndrome. Nat Rev Cancer 2005;5:11-20. [PubMed]
- Hasle H, Clemmensen IH, Mikkelsen M. Risks of leukaemia and solid tumours in individuals with Down's syndrome. Lancet 2000;355:165-9. [PubMed]
- NSW Mothers and Babies 2010. Sydney: Centre for Epidemiology and Evidence, NSW Ministry of Health, 2012.
- Malinge S, Izraeli S, Crispino JD. Insights into the manifestations, outcomes, and mechanisms of leukemogenesis in Down syndrome. Blood 2009;113:2619-28. [PubMed]
- Hasle H, Niemeyer CM, Chessells JM, et al. A pediatric approach to the WHO classification of myelodysplastic and myeloproliferative diseases. Leukemia 2003;17:277-82. [PubMed]
- Marshall GM, Carter DR, Cheung BB, et al. The prenatal origins of cancer. Nat Rev Cancer 2014;14:277-89. [PubMed]
- Schifferli A, Hitzler J, Bartholdi D, et al. Transient myeloproliferative disorder in neonates without Down syndrome: case report and review. Eur J Haematol 2015;94:456-62. [PubMed]
- Roy A, Roberts I, Norton A, et al. Acute megakaryoblastic leukaemia (AMKL) and transient myeloproliferative disorder (TMD) in Down syndrome: a multi-step model of myeloid leukaemogenesis. Br J Haematol 2009;147:3-12. [PubMed]
- Gamis AS, Smith FO. Transient myeloproliferative disorder in children with Down syndrome: clarity to this enigmatic disorder. Br J Haematol 2012;159:277-87. [PubMed]
- Haemmerling S, Behnisch W, Doerks T, et al. A 15q24 microdeletion in transient myeloproliferative disease (TMD) and acute megakaryoblastic leukaemia (AMKL) implicates PML and SUMO3 in the leukaemogenesis of TMD/AMKL. Br J Haematol 2012;157:180-7. [PubMed]
- Klusmann JH, Creutzig U, Zimmermann M, et al. Treatment and prognostic impact of transient leukemia in neonates with Down syndrome. Blood 2008;111:2991-8. [PubMed]
- Massey GV, Zipursky A, Chang MN, et al. A prospective study of the natural history of transient leukemia (TL) in neonates with Down syndrome (DS): Children's Oncology Group (COG) study POG-9481. Blood 2006;107:4606-13. [PubMed]
- Gamis AS, Alonzo TA, Gerbing RB, et al. Natural history of transient myeloproliferative disorder clinically diagnosed in Down syndrome neonates: a report from the Children's Oncology Group Study A2971. Blood 2011;118:6752-9. [PubMed]
- Roberts I, Alford K, Hall G, et al. GATA1-mutant clones are frequent and often unsuspected in babies with Down syndrome: identification of a population at risk of leukemia. Blood 2013;122:3908-17. [PubMed]
- Pine SR, Guo Q, Yin C, et al. Incidence and clinical implications of GATA1 mutations in newborns with Down syndrome. Blood 2007;110:2128-31. [PubMed]
- Henry E, Walker D, Wiedmeier SE, et al. Hematological abnormalities during the first week of life among neonates with Down syndrome: data from a multihospital healthcare system. Am J Med Genet A 2007;143A:42-50. [PubMed]
- Bull MJ. Committee on Genetics. Health supervision for children with Down syndrome. Pediatrics 2011;128:393-406. [PubMed]
- Lange BJ, Kobrinsky N, Barnard DR, et al. Distinctive demography, biology, and outcome of acute myeloid leukemia and myelodysplastic syndrome in children with Down syndrome: Children's Cancer Group Studies 2861 and 2891. Blood 1998;91:608-15. [PubMed]
- Sorrell AD, Alonzo TA, Hilden JM, et al. Favorable survival maintained in children who have myeloid leukemia associated with Down syndrome using reduced-dose chemotherapy on Children's Oncology Group trial A2971: a report from the Children's Oncology Group. Cancer 2012;118:4806-14. [PubMed]
- Taub JW, Huang X, Matherly LH, et al. Expression of chromosome 21-localized genes in acute myeloid leukemia: differences between Down syndrome and non-Down syndrome blast cells and relationship to in vitro sensitivity to cytosine arabinoside and daunorubicin. Blood 1999;94:1393-400. [PubMed]
- Ge Y, Stout ML, Tatman DA, et al. GATA1, cytidine deaminase, and the high cure rate of Down syndrome children with acute megakaryocytic leukemia. J Natl Cancer Inst 2005;97:226-31. [PubMed]
- Hasle H, Abrahamsson J, Arola M, et al. Myeloid leukemia in children 4 years or older with Down syndrome often lacks GATA1 mutation and cytogenetics and risk of relapse are more akin to sporadic AML. Leukemia 2008;22:1428-30. [PubMed]
- Roy A, Cowan G, Mead AJ, et al. Perturbation of fetal liver hematopoietic stem and progenitor cell development by trisomy 21. Proc Natl Acad Sci U S A 2012;109:17579-84. [PubMed]
- Maclean GA, Menne TF, Guo G, et al. Altered hematopoiesis in trisomy 21 as revealed through in vitro differentiation of isogenic human pluripotent cells. Proc Natl Acad Sci U S A 2012;109:17567-72. [PubMed]
- Tunstall-Pedoe O, Roy A, Karadimitris A, et al. Abnormalities in the myeloid progenitor compartment in Down syndrome fetal liver precede acquisition of GATA1 mutations. Blood 2008;112:4507-11. [PubMed]
- Wechsler J, Greene M, McDevitt MA, et al. Acquired mutations in GATA1 in the megakaryoblastic leukemia of Down syndrome. Nat Genet 2002;32:148-52. [PubMed]
- Rainis L, Bercovich D, Strehl S, et al. Mutations in exon 2 of GATA1 are early events in megakaryocytic malignancies associated with trisomy 21. Blood 2003;102:981-6. [PubMed]
- Li Z, Godinho FJ, Klusmann JH, et al. Developmental stage-selective effect of somatically mutated leukemogenic transcription factor GATA1. Nat Genet 2005;37:613-9. [PubMed]
- Sankaran VG, Ghazvinian R, Do R, et al. Exome sequencing identifies GATA1 mutations resulting in Diamond-Blackfan anemia. J Clin Invest 2012;122:2439-43. [PubMed]
- Hollanda LM, Lima CS, Cunha AF, et al. An inherited mutation leading to production of only the short isoform of GATA-1 is associated with impaired erythropoiesis. Nat Genet 2006;38:807-12. [PubMed]
- Hitzler JK, Cheung J, Li Y, et al. GATA1 mutations in transient leukemia and acute megakaryoblastic leukemia of Down syndrome. Blood 2003;101:4301-4. [PubMed]
- Alford KA, Reinhardt K, Garnett C, et al. Analysis of GATA1 mutations in Down syndrome transient myeloproliferative disorder and myeloid leukemia. Blood 2011;118:2222-38. [PubMed]
- Yoshida K, Toki T, Okuno Y, et al. The landscape of somatic mutations in Down syndrome-related myeloid disorders. Nat Genet 2013;45:1293-9. [PubMed]
- Nikolaev SI, Santoni F, Vannier A, et al. Exome sequencing identifies putative drivers of progression of transient myeloproliferative disorder to AMKL in infants with Down syndrome. Blood 2013;122:554-61. [PubMed]
- Queiroz LB, Lima BD, Mazzeu JF, et al. Analysis of GATA1 mutations and leukemogenesis in newborns with Down syndrome. Genet Mol Res 2013;12:4630-8. [PubMed]
- Salek-Ardakani S, Smooha G, de Boer J, et al. ERG is a megakaryocytic oncogene. Cancer Res 2009;69:4665-73. [PubMed]
- Stankiewicz MJ, Crispino JD. ETS2 and ERG promote megakaryopoiesis and synergize with alterations in GATA-1 to immortalize hematopoietic progenitor cells. Blood 2009;113:3337-47. [PubMed]
- Birger Y, Goldberg L, Chlon TM, et al. Perturbation of fetal hematopoiesis in a mouse model of Down syndrome's transient myeloproliferative disorder. Blood 2013;122:988-98. [PubMed]
- Ng AP, Hyland CD, Metcalf D, et al. Trisomy of Erg is required for myeloproliferation in a mouse model of Down syndrome. Blood 2010;115:3966-9. [PubMed]
- Malinge S, Bliss-Moreau M, Kirsammer G, et al. Increased dosage of the chromosome 21 ortholog Dyrk1a promotes megakaryoblastic leukemia in a murine model of Down syndrome. J Clin Invest 2012;122:948-62. [PubMed]
- Rainis L, Toki T, Pimanda JE, et al. The proto-oncogene ERG in megakaryoblastic leukemias. Cancer Res 2005;65:7596-602. [PubMed]
- Elagib KE, Racke FK, Mogass M, et al. RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood 2003;101:4333-41. [PubMed]
- Kirsammer G, Jilani S, Liu H, et al. Highly penetrant myeloproliferative disease in the Ts65Dn mouse model of Down syndrome. Blood 2008;111:767-75. [PubMed]
- Song WJ, Sullivan MG, Legare RD, et al. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet 1999;23:166-75. [PubMed]
- Chou ST, Byrska-Bishop M, Tober JM, et al. Trisomy 21-associated defects in human primitive hematopoiesis revealed through induced pluripotent stem cells. Proc Natl Acad Sci U S A 2012;109:17573-8. [PubMed]
- Bourquin JP, Subramanian A, Langebrake C, et al. Identification of distinct molecular phenotypes in acute megakaryoblastic leukemia by gene expression profiling. Proc Natl Acad Sci U S A 2006;103:3339-44. [PubMed]
- McElwaine S, Mulligan C, Groet J, et al. Microarray transcript profiling distinguishes the transient from the acute type of megakaryoblastic leukaemia (M7) in Down's syndrome, revealing PRAME as a specific discriminating marker. Br J Haematol 2004;125:729-42. [PubMed]
- Bercovich D, Ganmore I, Scott LM, et al. Mutations of JAK2 in acute lymphoblastic leukaemias associated with Down's syndrome. Lancet 2008;372:1484-92. [PubMed]
- Letourneau A, Santoni FA, Bonilla X, et al. Domains of genome-wide gene expression dysregulation in Down's syndrome. Nature 2014;508:345-50. [PubMed]
- Toki T, Kanezaki R, Adachi S, et al. The key role of stem cell factor/KIT signaling in the proliferation of blast cells from Down syndrome-related leukemia. Leukemia 2009;23:95-103. [PubMed]
- Liu L, Wen Q, Gong R, et al. PSTPIP2 dysregulation contributes to aberrant terminal differentiation in GATA-1-deficient megakaryocytes by activating LYN. Cell death & disease 2014;5:e988. [PubMed]
- Klusmann JH, Li Z, Böhmer K, et al. miR-125b-2 is a potential oncomiR on human chromosome 21 in megakaryoblastic leukemia. Genes Dev 2010;24:478-90. [PubMed]
- Shaham L, Vendramini E, Ge Y, et al. MicroRNA-486-5p is an erythroid oncomiR of the myeloid leukemias of Down syndrome. Blood 2015;125:1292-301. [PubMed]
- Woo AJ, Wieland K, Huang H, et al. Developmental differences in IFN signaling affect GATA1s-induced megakaryocyte hyperproliferation. J Clin Invest 2013;123:3292-304.
- Tsuzuki S, Taguchi O, Seto M. Promotion and maintenance of leukemia by ERG. Blood 2011;117:3858-68. [PubMed]
- Carmichael CL, Majewski IJ, Alexander WS, et al. Hematopoietic defects in the Ts1Cje mouse model of Down syndrome. Blood 2009;113:1929-37. [PubMed]
- Alford KA, Slender A, Vanes L, et al. Perturbed hematopoiesis in the Tc1 mouse model of Down syndrome. Blood 2010;115:2928-37. [PubMed]
- De Vita S, Canzonetta C, Mulligan C, et al. Trisomic dose of several chromosome 21 genes perturbs haematopoietic stem and progenitor cell differentiation in Down's syndrome. Oncogene 2010;29:6102-14. [PubMed]
- Wen Q, Goldenson B, Silver SJ, et al. Identification of regulators of polyploidization presents therapeutic targets for treatment of AMKL. Cell 2012;150:575-89. [PubMed]
- Saida S, Watanabe K, Sato-Otsubo A, et al. Clonal selection in xenografted TAM recapitulates the evolutionary process of myeloid leukemia in Down syndrome. Blood 2013;121:4377-87. [PubMed]
- Lane AA, Chapuy B, Lin CY, et al. Triplication of a 21q22 region contributes to B cell transformation through HMGN1 overexpression and loss of histone H3 Lys27 trimethylation. Nat Genet 2014;46:618-23. [PubMed]
- Khan I, Malinge S, Crispino J. Myeloid leukemia in Down syndrome. Crit Rev Oncog 2011;16:25-36. [PubMed]
- Kaneko H, Kobayashi E, Yamamoto M, et al. N- and C-terminal transactivation domains of GATA1 protein coordinate hematopoietic program. J Biol Chem 2012;287:21439-49. [PubMed]
- Ahmed M, Sternberg A, Hall G, et al. Natural history of GATA1 mutations in Down syndrome. Blood 2004;103:2480-9. [PubMed]
- Groet J, McElwaine S, Spinelli M, et al. Acquired mutations in GATA1 in neonates with Down's syndrome with transient myeloid disorder. Lancet 2003;361:1617-20. [PubMed]
- Kanezaki R, Toki T, Terui K, et al. Down syndrome and GATA1 mutations in transient abnormal myeloproliferative disorder: mutation classes correlate with progression to myeloid leukemia. Blood 2010;116:4631-8. [PubMed]
- Caldwell JT, Edwards H, Buck SA, et al. Targeting the wee1 kinase for treatment of pediatric Down syndrome acute myeloid leukemia. Pediatr Blood Cancer 2014;61:1767-73. [PubMed]
- Caldwell JT, Edwards H, Dombkowski AA, et al. Overexpression of GATA1 confers resistance to chemotherapy in acute megakaryocytic Leukemia. PloS one 2013;8:e68601. [PubMed]
- Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood 2009;114:937-51. [PubMed]
- Mateos MK, O'Brien TA, Oswald C, et al. Transplant-related mortality following allogeneic hematopoeitic stem cell transplantation for pediatric acute lymphoblastic leukemia: 25-year retrospective review. Pediatr Blood Cancer 2013;60:1520-7. [PubMed]
- Buitenkamp TD, Izraeli S, Zimmermann M, et al. Acute lymphoblastic leukemia in children with Down syndrome: a retrospective analysis from the Ponte di Legno study group. Blood 2014;123:70-7. [PubMed]
- Hertzberg L, Vendramini E, Ganmore I, et al. Down syndrome acute lymphoblastic leukemia, a highly heterogeneous disease in which aberrant expression of CRLF2 is associated with mutated JAK2: a report from the International BFM Study Group. Blood 2010;115:1006-17. [PubMed]
- Nikolaev SI, Garieri M, Santoni F, et al. Frequent cases of RAS-mutated Down syndrome acute lymphoblastic leukaemia lack JAK2 mutations. Nature communications 2014;5:4654. [PubMed]
- Izraeli S, Vora A, Zwaan CM, et al. How I treat ALL in Down's syndrome: pathobiology and management. Blood 2014;123:35-40. [PubMed]
- Patrick K, Wade R, Goulden N, et al. Outcome of Down syndrome associated acute lymphoblastic leukaemia treated on a contemporary protocol. Br J Haematol 2014;165:552-5. [PubMed]
- Taub JW, Ge Y. Down syndrome, drug metabolism and chromosome 21. Pediatr Blood Cancer. 2005;44:33-9. [PubMed]
- Cervantes F, Vannucchi AM, Kiladjian JJ, et al. Three-year efficacy, safety, and survival findings from COMFORT-II, a phase 3 study comparing ruxolitinib with best available therapy for myelofibrosis. Blood 2013;122:4047-53. [PubMed]
- Roberts KG, Pei D, Campana D, et al. Outcomes of children with BCR-ABL1–like acute lymphoblastic leukemia treated with risk-directed therapy based on the levels of minimal residual disease. J Clin Oncol 2014;32:3012-20. [PubMed]
- Hong D, Gupta R, Ancliff P, et al. Initiating and cancer-propagating cells in TEL-AML1-associated childhood leukemia. Science 2008;319:336-9. [PubMed]
- Gefen N, Binder V, Zaliova M, et al. Hsa-mir-125b-2 is highly expressed in childhood ETV6/RUNX1 (TEL/AML1) leukemias and confers survival advantage to growth inhibitory signals independent of p53. Leukemia 2010;24:89-96. [PubMed]
- Forestier E, Izraeli S, Beverloo B, et al. Cytogenetic features of acute lymphoblastic and myeloid leukemias in pediatric patients with Down syndrome: an iBFM-SG study. Blood 2008;111:1575-83. [PubMed]
- Malinge S, Chlon T, Dore LC, Ketterling RP, Tallman MS, Paietta E, et al. Development of acute megakaryoblastic leukemia in Down syndrome is associated with sequential epigenetic changes. Blood 2013;122:e33-43. [PubMed]


