Spanning the genomics era: the vital role of a single institution biorepository for childhood cancer research over a decade
Introduction: biospecimens and the changing nature of translational genomics
It is considered that understanding the genetic basis of human disease will guide translational research and determine future patient treatment courses. For over a decade translational research has shifted from genetics, the study of individual genes in an organism, to genomics, the study of the complete set of genes within a single organism (1). Cancer is a genetic disorder. Over the past two decades, our understanding of genomics has begun to directly impact the way we diagnose, risk stratify and treat cancer. Before the genomic era, cancer was classified by site of the origin in the body. Now it is by the prototypic surface markers, gross karyotypic changes and selected DNA/RNA markers (2). Consequently, novel, less toxic and personally directed treatment strategies have been designed that target the specific genetic alternations for the cancer (3). Furthermore, the widespread use of genomic technologies, in particular next generation sequencing, has dramatically enhances the understanding of drug resistance and offer rationales for combination therapies (2).
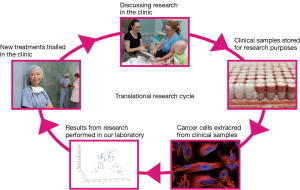
The ‘genomics era’ is considered to have begun with the commencement of the Human Genome Project when advancements in technology led to high-through-put sequencing of DNA and RNA. With its promise of personalised medicine the focus on the need for human tissue within our research culture has never been so heightened. Indeed, translational genomic studies can only be established when human tissue samples are available for analysis. This inextricable requirement for tissue during the genomics era led to the growing interdependence between genomics researchers and clinical practitioners. This two-way or cyclical relationship between domain experts in the sequencing laboratories and the medical clinics now underpin the translational potential of the technological developments in genomics (Figure 1).
Biospecimens are now proven to be essential elements for successful translational genomics research especially within this relationship. Not only does the biospecimen contain the genetic material to be sequenced by the genomic researcher, but when put in context with the patient pathological and clinical records creates a rich data source with which to understand a patients disease. Hence, the systematic collection of tissue samples into ‘biobanks’ has become a fundamental activity in many medico-research centres (4). The development of biobanking as a standalone scientific discipline is well advanced yielding a new population of professionals specifically experienced in governance, regulation, ethics and values, law, project management, marketing, business, science and all pathology disciplines required for tissue resourcing.
During the genomics era the necessity for more extensive biobanking infrastructure and operations has emerged (5). A strong theme in the biobanking literature is as “there is a need for large numbers of samples to provide statistical power to research studies….a single biobank cannot provide sufficient numbers of samples to capture the full spectrum of any disease.” (6). As translational genomic research became an international collaborative effort, broad based national and international biobanking networks have become common place (7). For example, in 2010, the Pediatric Cancer Genome Project was launched by St. Jude Children’s Research Hospital and The Genome Institute at Washington University, with the goal of sequencing the whole genome of 600 paediatric tumours and matched non-tumour samples (3). It is strongly anticipated that data from such extensive investigations will dramatically enhance the understanding of paediatric cancer and affect our clinical management of patients. Similarly, the International Cancer Genome Consortium (ICGC) established standardised biobanking practice as an initial phase of its strategies to which adherence was mandatory for inclusion in the wider sequencing project (8).
How are current genomic studies affecting biobanking?
As genetic studies exploring single ‘candidate’ genes or undertaking step-by-step approaches like positional cloning make way for high-through-put multiplex PCR, microarray based techniques or one of the wide range of next generation sequencing platforms, the ability to generate massive amounts of genomic data in now occurring in rapid time. This requires different analytical strategies and experimental designs. Rather than studying one genetic region in depth on the basis of a hypothesis driven strategy, the high-through-put techniques associated with current genomic studies has led to ‘screening’ based experiments with technology-driven generation of the data and the subsequent exploration of the data for interesting patterns and trends and for disease ‘biomarkers’.
That genomic studies generate large amounts of data has presented novel issues with the design of these studies which has consequential effects on biobanking pathways. The ‘curse of dimensionality’ (9), where the number of data points gathered for each sample far outweigh the number of samples available leads to a high a high false positive rate. The immediate solution is for the establishment of national and international biobanking networks and the systematic collection of large numbers of tissue. However, with genomics yielding millions of polymorphisms per sample it is difficult to see how even this can yield the numbers required to meet current power requirements. Further, genomic complexity impedes the ability for data models to generalize across sample cohorts sufficiently well. For genomic markers found during analysis to be validated using downstream techniques requires ample samples to be stored in different formats (e.g., frozen, formalin-fixed, fresh) to allow the full spectrum of analysis for protein products as well as genes.
At the commencement of the genomics era, study design aimed to identify key features in simple, more defined and easily controlled systems like cell lines. These features would then be explored in biospecimens to see if the candidate gene was present within the tissue. With the genomic era this was reversed. Biospecimens are no longer seen as the environment to test whether the researcher’s candidate gene data is meaningful. Rather, translational genomics see the biospecimen itself as the data. This data is now easily extracted, deciphered and organised for further interrogation which often takes place in defined experimental models like cell line or animals. Moreover, there is also an imperative for the biospecimen to be linked to a full range of patient clinical data and their clinical treatment so that the data can be interpreted with a translational intent. With the genomics era, the main ‘tool’ used by the biobanker is not the freezer, but the database. The value of a biobank is not just based on sample holdings, but on diagnostic, demographic, tissue processing metrics and clinical data collected that is subsequently associated with the samples and indeed the patients. To this end, tissue sample can be viewed as ‘little packages of information’ making biobanking as much an informatics problem as a pathology problem. The true value of a biobanks is bound up in the information they supply, and should be acting as ‘hubs’ of valuable data, requiring intricate IT backbones to support this activity. Unfortunately many biobanks rely on a piecemeal database systems, generally capturing information about the tissue specimens in a manner removed from the patient records. Regardless, advances in genomics have highlighted the data potential of biospecimens and the value of systematic biobanking activity to translational research.
How should biobanking affect current genomic studies?
The advancement of biobanks as specialist research facilities is evidence of the growing recognition that stores of human tissue provide enticing opportunities for researcher to explore their new discoveries in light of potential clinical applications. It is difficult to say whether the growth in the prevalence of biobanking has been driven out of a need to translate our research findings into clinical practice, or whether the increased motivation for medical researchers to undertake translational investigations has been driven by the opportunity forded to them by the establishment of biobanks. As biobanking is becoming integral within clinical services and trials, we are seeing that, in essence, the practice of biobanking is causing a culture change within our clinical institutions. Translational research is becoming more and more an extension on patient healthcare practices, and provides greater opportunity for clinicians to learn from past patient experience and derive new knowledge which will influence how the next cohort of patients will be treated.
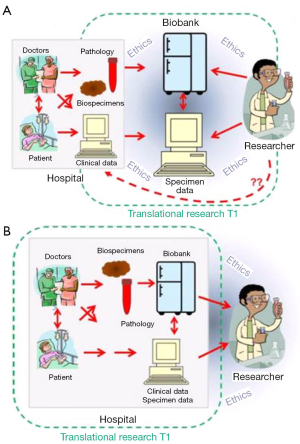
Biobanking emerged as a practice built upon investigator-driven activity, essential due to researchers need for quality tissue to answer their specific questions. In this situation ‘pull out biobanking’ where biospecimens are retrieved from within clinical services and transferred to a specialist ‘biorepository’, which are often placed within the research centres or an off-site centralized facility, away from the immediate collection source (pathology department, medical clinic). It is recognised that this environment does not always lend itself to clear and greater access to tissue specimens (10), in particular as hospital ethics boards need to approve and monitor the biorepository as well as the research initiatives it supports (Figure 2A). Through the genomics era however, we have seen a call to ‘push out biobanking’, the institution-driven activity where biobanks operate as an extension on routine hospital practice that leverages established pathways to ensure tissue samples and related clinical data are accessible for research projects. Biobanks within this model are built on standardized practices, are patient focused and require the health providers to enable biobanks within their local sites but to engage with researchers to ensure appropriate and ethical use of tissue in research. Further, the specimen data about tissue processing, storage condition and researcher access that would be stored within a biobank becomes part of the patient record (Figure 2B). In doing so, throughout the genomics era, biobanking has allowed the potential of the genomic information within the biospecimens to be explored and understood directly in the patient context.
Translational genomics for childhood cancer: the rare diseases paradigm
With the increased number of genomic studies into cancer, it is considered that the availability of biospecimens will become the rate limiting step (11). Despite the efforts in international biobanking, translational genomics is hampered when there low numbers of biospecimens for a particular disease and there are necessary long collection times required. This is most apparent for paediatric cancer as it is considered a rare disease. A ‘rare disease’ is defined by (12) as having a prevalence 500 per million and often manifest in newborns or children. The clinical burden of rare disease is often exacerbated by a lack of specific treatment for patients and, with the prevalence of paediatric cancer in Australia being 160 cases per million for all cancer types, this is never a more relevant paradigm. Childhood cancer is the leading cause of disease-related death among children and adolescents (ages 1 to 19 years) (13). Worldwide, the estimated incidence rate is more than 175,000 per year and the mortality rate varies from 20% (in developed countries) to 80% (in developing countries). Paediatric cancers are very different from adult’s cancers in type, underlying pathogenesis, biology, response rate and outcome. Paediatric cancers are mainly embryonic in origin and are often the result of DNA changes in cells during the early stage of life.
When addressing the need for tumour biospecimen procurement to support translational genomics into paediatric cancer, researchers are left with two main options. Firstly, researchers are required to perform worldwide sweeps of material that may have already been collected. This will require international collaboration and data integration from different centres to achieve the critical mass required (14). In this situation the challenge for the experimenter is negotiating the different collection practices of biobanks and ensuring that any differences doesn’t introduce a bias or confounder to upstream data analysis. Capturing consistent sample-matched clinical data across multiple centres prove the most difficult as there is considerable variance in records management practices as well as different stringencies around local privacy regulations.
Secondly, solutions have been sought through networking of biobanks that have a common function and purpose (6,15,16). Through networking, the promotion, optimization and dissemination of quality material and associated data to scientific users as well as biobank-pertinent knowledge and know-how would meet the collective needs of translational genomics researchers. The challenge is to ensure that biobank linkage or interoperability is seamless and scalable as the network grows. The difficulty is the need for extra layers of regulation, management and infrastructure that are required to allow networked biobanks to communicate and function together.
Keeping it simple: the role of a single institution biorepository
Clearly, there is a call for biobanking practice to be responsive to the current experimental needs of the time and more expansive systems for tissue procurement are being called for to meet international demands (14). The current dialogue poses that translational research is dependent on ‘harmonized’ biobanking practices that function as a major system within national and international research agendas.
“We are moving to a future where biobanks, existing biorepositories and reference databases will be linked and networked for research purposes in ways that has not been possible before.” (6).
“There is a growing recognition that a common strategy is imperative to develop biobanking globally and effectively.….fuelling opportunities for biobanks to ultimately become integral parts of health-care systems in many countries.” (7).
In this ‘brave new world’ of international biobanking superstructures, what can be learned from the past experiences of local biobanks? Do institutional biorepositories have a place in the modern translational genomics, or should they all be superseded into major networking infrastructure or larger collaborative efforts for them to have an impact? If this is truly the case, what role does a single institution biorepository play in the current highly networked world of translational genomics. Here we describe such a case.
The Tumour Bank at The Children’s Hospital at Westmead (TB-CHW) in the western suburbs of Sydney was formally established in 1998 with the financial assistance of benefactor funds donated to the Oncology Department to support research. Operating as a single institutional biobank within The Children’s Hospital at Westmead, the TB-CHW is a biorepository that co-ordinates the collection, storage and distribution of paediatric biospecimens including paraffin-embedded formalin fixed tissue, fresh frozen tumours, bone marrow aspirates, cerebrospinal fluid and peripheral blood samples donated by patients and obtained through the normal clinical management pathways operating through the hospital. The TB-CHW follows an open access policy and is available to all researchers. Specimens are made available to researchers from around the world through an application process that is overseen by the institutional Tumour Bank Committee, Human Research Ethics Committee and Governance Department. Further, related tissue handling expertise is provides by TB-CHW technical personnel including pathology validation with cellular, genomic, and proteomic preparation and quality assurance analysis. Customized collection and processing of tissue for prospective studies or clinical trials is also supported. Consequently, as a key resource for translational paediatric cancer research, the TB-CHW has developed into a vital biospecimen repository for the cancer research community.
The TB-CHW has been operating throughout the genomics era during which time it has increasingly found itself having a vital role in the establishment of translational genomics for paediatric cancer. It needs to be born in mind that the decision to collect samples into the TB-CHW precedes the research questions being asked. At the TB-CHW, samples are not collected for specific studies but rather during the course of patient management as a routine activity. Staff, embedded within the institution, follow standard operating procedures derived in-house and refined over time. The experience gained by the TB-CHW in meeting the needs of researchers during this period has been informative to the present calls for expansive biobanking.
Here we detail metrics that demonstrate how a single institution biorepository, in its own right, has been a strong participant in the advancement of translational genomics throughout the genomics era. We demonstrate that simple biobanking practices, when performed well, are often all that are required to meet the needs of major efforts with genomics and will enable the advancements expected for translational research. Furthermore, we illustrate how a rare diseases biobank can facilitate the inclusion of small groups of patients into significant international studies conducted within the major research centres around the world. For our evaluation we considered research to be included as any project conducted within the last 15 years that investigated studies investigating single genes, genetic signatures, whole genome data or epigenetic markers (e.g., methylation). As there is still no universally accepted means to measure biobanks contribution to research (17), we have gauged the success if the TB-CHW with these studies using a simple work flow process: (I) the specimen distribution including numbers of samples; (II) where the research were conducted and what studies did they contributed to; (III) were the outcomes published and what was the quality of the publication and did the knowledge gained lead to further research, contribute to clinical trials or change in patient management practice.
Contribution to research: national and international distribution profile
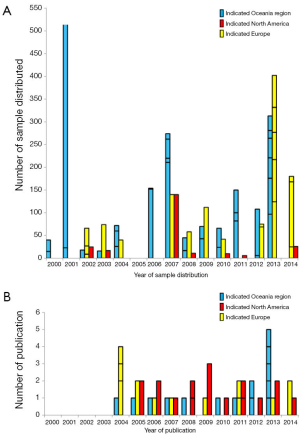
The TB-CHW has supported a total 84 projects undertaken by researchers located around the world. Among them more than 68% (n=57) were considered as gene or genomic based studies. Studies considered for further discussion involved techniques used to explore single genes (PCR), genetic signatures (multiplex PCR, microarray), whole genome (next generation sequencing) and epigenetics (e.g., methylation). The TB-CHW did not experience a strong ‘ripple effect’ of sample distribution from local researchers to regional to national and then international users (Figure 3A). Indeed, primary centres from the USA and Europe were some of the initial users of the TB-CHW as it was gaining momentum locally and throughout Australia. The TB-CHW does not have a passive marketing approach. Rather its mandate is to actively promote our collection to leading researchers whose investigations are deemed likely to benefit from inclusion of the specimens we have in store and whose results will benefit the broader understanding of paediatric malignancy. Through this direct approach, the TB-CHW has been active contributors to a range of leading research activities performed in institution from around the world (Figure 3, Table 1).
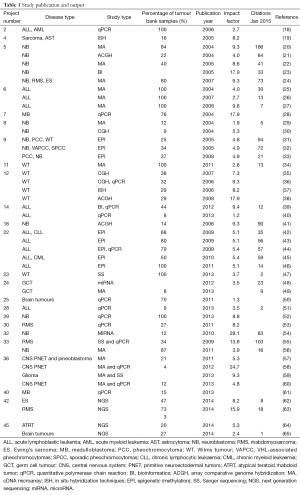
Full table
Within the changing nature of translational research, the TB-CHW has supported study designs that have shifted from gene based assessments to whole-genome interrogations (Figure 4). Since the commencement of our activity, gene based research was restricted to the validation of the role of candidate genes or small gene families. With the genomics era commencing a shift in focus emerged with researchers performing microarray based studies seeking samples. This led to more exploratory, less hypothesis-driven studies that delved into gene expression data. At this point biobanked primary tumour that had been flash-frozen in liquid nitrogen were in high demand partly due to the nature of the technology as much as the research questions being asked. As microarray technology advanced applications were developed that assessed DNA sequence polymorphism and germ-line variation to understand heritable differences in patients, applications received by the TB-CHW similarly shifted to requiring ‘normal’ samples. Peripheral blood collected by the TB-CHW as part of its routine practice then became highly sought after specimens. Further refinement of the technologies designed to interrogate deeper levels of genomic regulation including miRNA screening and epigenetics (e.g., methylation) requests returned to requiring primary tumours. Throughout these periods, most studies were adequately supplied through the TB-CHW with samples collected retrospectively. In few cases ongoing collection and prospective provision of samples to a study was required.
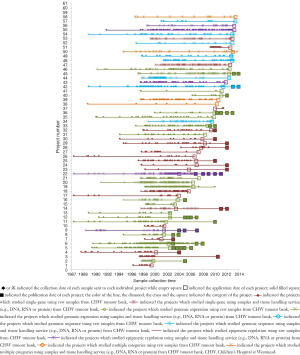
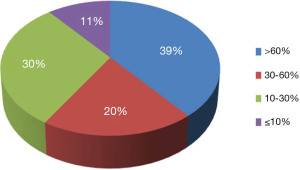
To demonstrate the TB-CHW ability to provide a consistent distribution of tissue throughout the genomics era, Figure 4 shows the sample collection time lines for each study supported. It can been seen that for most studies, sample provided had been collected over an extended period with some cases, principally those using paraffin embedded tissue sourced from the hospitals pathology archive, collections from a 20 year period or more being supplied. The average collection period is 138±69 months or over a decade. At the extremes, only 3 (5%) studies utilised samples collected over a period of no more than 24 months whilst 17 studies (27%) received samples collected over a period of 15 years or more (Figure 4). This range of distribution did not diminish as more studies were approved reflecting a consistency in the TB-CHW ability to meet ‘supply and demand’ needs of researchers. Nor was the range of sample provision influenced by the type of study being performed, whether it would be single gene validation, whole genome or exome screening. This result needs to be considered in light of recently espoused best practices for biospecimen handling for genomics studies (66) where being compliant to these standards was outweighed by the overwhelming need of the researcher to obtain enough samples for their investigation regardless of whether they were collected before the best practices were enforced or not. Researchers applying to the TB-CHW were not concerned about mixing samples that may have been likely collected following differing protocols. Over 60% of the samples provided were studied alongside specimens collected from multiple sites. For 11% of the studies supported (projects 14, 22, 26, 34), the TB-CHW contribution to those studies was less than 10% of the total collection which reflects the investigators undertaking a world-wide sweep for a particular very rare tumour (Figure 5).
Contribution to knowledge: locals gaining international profiles
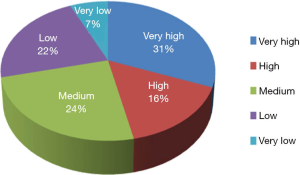
The TB-CHW has contributed to an outstanding scientific output in paediatric genomic studies (Table 1). The TB-CHW publication record directly mirrors the advancement of genomics and related technology. More than 40 genomic publications have been generated from those projects. Publication quality has been distributed evenly within broad categories. Figure 6 shows the percentage of publications according to their impact factors assembled into broad ranges. The average impact factor is 7.58 but over 75% of the publications have impact factors greater than 5. The publications have been generated in multiple disease categories, most from neuroblastoma, followed by leukemia, Wilm’s tumour (Table 1). International researchers have contributed 76.2% publications while only 23.8% are from local researchers exemplifying the extent of the contribution of the TB-CHW to world-wide efforts in paediatric cancer research. The number of publications gained per year correlate with the projects supported indicating a consistency in the TB-CHW’s long term contribution to the knowledge gained through these research endeavours (Figure 3B).
Finally, as The Children’s Hospital at Westmead is dedicated to paediatric healthcare, it provides services to children and their families in the wider western metropolitan areas of Sydney and regional centres. It runs one of two major Oncology Treatment Centres for New South Wales and admits up to 150 new cancer patients each year. Of most significance, through the activities of TB-CHW this small population of patients have been entered into world class, cutting edge research programs performed in leading research centres around the world. The TB-CHW has allowed us to gain knowledge about our local patients whilst comparing them with patients from other lands with similar disorder. In so doing the relevance of published research to our local patients is immediately and directly known. Moreover, the opportunity for the hospital to benefit from this knowledge through new treatment options is enhanced as the clinical history of the patients can be directly linked to the data derived from each study.
Concluding statement
This paper describes the significant contribution of a single institutional hospital embedded tumour biobank to the genomic research community, especially for rare diseases. Despite the increased stringencies placed on biobanking practice, the TB-CHW has shown that a single institution biorespository can have a consistent and effective contribution to translational research into paediatric malignancy demonstrating its long term benefit throughout the genomics era. This is significant in that patient samples from rare diseases like paediatric cancer have a relative higher value as it will take longer time and more effort to accumulate enough samples to gain the statistical power. For rare disease research, every sample counts! Hence, the vision of the TB-CHW to establish routine tissue collection strategies for research over 15 years ago has brought it to the forefront of the advances in genomics that have advanced throughout this period. The TB-CHW experience demonstrates that the success and impact of a single institution biorepository lies in its proactivity. The potential of many hospital institutions to contribute to the global research efforts through provision of tissue samples and linked clinical data are inhibited by biobanking notions that a specialist facility is required. Such a facility is required to adhere to highly specific criteria that meet best practice, answer specific research questions in their own right, must be linked to other partners in a biobanking network through common processes, be technologically supported with specialist equipment and software, is required to be stringently regulated by health governance bodies and consequently requires lots of funding (67,68). Whilst the TB-CHW endorses and strives for these high ideals in biobanking, having actively contributed to the formation of the national Australasian Biospecimens Network Association (http://abrn.net), it has been more keenly driven by the greater need that research must be done on rare diseases, like paediatric cancer, for us to improve health outcomes for these patients. The success of the TB-CHW through-out the genomics era has therefore been driven by three related principles.
Embedded biobanking
Biospecimen procurement for research must occur within the clinical pathways of the institution as a routine daily procedure. In this sense the practice of biobanking precedes the research questions to be addressed. This creates the flexibility within the institution to address all types of research projects.
Exhaustive biobanking
With embedded biobanking the opportunity exists to procure every biospecimen through common mechanisms within our institution. This allows for the comprehensive collection of all samples from rare diseases, over lengthy periods of time if required. This creates the motivation for the institution to practically support all types of research projects.
Engaging biobanking
Having a comprehensive collection of tissue for a rare disease allows for its biobankers to actively engage with the wider scientific community through open access biobanking policies. ‘Push out’ biobanking practices aid to remove barriers and allow the institution to actively participate in the leading genomic research efforts ongoing around the world. This creates the opportunity for the institution to participate in all types of research projects.
Acknowledgements
The Tumour Bank at The Children’s Hospital at Westmead wishes to acknowledge the ongoing funding provided by The Kids Cancer Project and past support from Kayaking for Chemo Kids, The Miles-2-Go Foundation and the Jenolan Caves Reserve Trust. The Tumour Bank has also been financed through Australia’s National Health and Medical Research Councils Enabling Grant scheme and with funds provided by Cancer Institute of New South Wales.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Offit K. Personalized medicine: new genomics, old lessons. Hum Genet 2011;130:3-14. [PubMed]
- Roychowdhury S, Chinnaiyan AM. Translating genomics for precision cancer medicine. Annu Rev Genomics Hum Genet 2014;15:395-415. [PubMed]
- Downing JR, Wilson RK, Zhang J, et al. The Pediatric Cancer Genome Project. Nat Genet 2012;44:619-22. [PubMed]
- Cole A, Cheah S, Dee S, et al. Biospecimen use correlates with emerging techniques in cancer research: impact on planning future biobanks. Biopreserv Biobank 2012;10:518-25. [PubMed]
- Cambon-Thomsen A. The social and ethical issues of post-genomic human biobanks. Nat Rev Genet 2004;5:866-73. [PubMed]
- Quinlan PR, Mistry G, Bullbeck H, et al. A data standard for sourcing fit-for-purpose biological samples in an integrated virtual network of biobanks. Biopreserv Biobank 2014;12:184-91. [PubMed]
- Harris JR, Burton P, Knoppers BM, et al. Toward a roadmap in global biobanking for health. Eur J Hum Genet 2012;20:1105-11. [PubMed]
- Esgueva R, Park K, Kim R, et al. Next-generation prostate cancer biobanking: toward a processing protocol amenable for the International Cancer Genome Consortium. Diagn Mol Pathol 2012;21:61-8. [PubMed]
- Catchpoole DR, Kennedy P, Skillicorn DB, et al. The curse of dimensionality: a blessing to personalized medicine. J Clin Oncol 2010;28:e723-4; author reply e725.
- Verlinden M, Nys H, Ectors N, et al. Access to biobanks: harmonization across biobank initiatives. Biopreserv Biobank 2014;12:415-22. [PubMed]
- Genomics Dancey J. personalized medicine and cancer practice. Clin Biochem 2012;45:379-81. [PubMed]
- Graham CE, Molster C, Baynam GS, et al. Current trends in biobanking for rare diseases: a review. Journal of Biorepository Science for Applied Medicine 2014;2:42.
- Youlden DR, Baade PD, Valery PC, et al. Childhood cancer mortality in Australia. Cancer Epidemiol 2012;36:476-80. [PubMed]
- Zatloukal K, Hainaut P. Human tissue biobanks as instruments for drug discovery and development: impact on personalized medicine. Biomark Med 2010;4:895-903. [PubMed]
- Kaye J. From single biobanks to international networks: developing e-governance. Hum Genet 2011;130:377-82. [PubMed]
- Roazzi P, Di Benedetto C, Bravo E, et al. Biobank networking: the European network initiative and the italian participation. Biopreserv Biobank 2011;9:175-9. [PubMed]
- Mabile L, Dalgleish R, Thorisson GA, et al. Quantifying the use of bioresources for promoting their sharing in scientific research. Gigascience 2013;2:7. [PubMed]
- Barbaric D, Byth K, Dalla-Pozza L, et al. Expression of tumor protein D52-like genes in childhood leukemia at diagnosis: clinical and sample considerations. Leuk Res 2006;30:1355-63. [PubMed]
- Henson JD, Hannay JA, McCarthy SW, et al. A robust assay for alternative lengthening of telomeres in tumors shows the significance of alternative lengthening of telomeres in sarcomas and astrocytomas. Clin Cancer Res 2005;11:217-25. [PubMed]
- Wei JS, Greer BT, Westermann F, et al. Prediction of clinical outcome using gene expression profiling and artificial neural networks for patients with neuroblastoma. Cancer Res 2004;64:6883-91. [PubMed]
- Chen QR, Bilke S, Wei JS, et al. cDNA array-CGH profiling identifies genomic alterations specific to stage and MYCN-amplification in neuroblastoma. BMC Genomics 2004;5:70. [PubMed]
- Krasnoselsky AL, Whiteford CC, Wei JS, et al. Altered expression of cell cycle genes distinguishes aggressive neuroblastoma. Oncogene 2005;24:1533-41. [PubMed]
- Bilke S, Chen QR, Westerman F, et al. Inferring a tumor progression model for neuroblastoma from genomic data. J Clin Oncol 2005;23:7322-31. [PubMed]
- Whiteford CC, Bilke S, Greer BT, et al. Credentialing preclinical pediatric xenograft models using gene expression and tissue microarray analysis. Cancer Res 2007;67:32-40. [PubMed]
- Mitchell SA, Brown KM, Henry MM, et al. Inter-platform comparability of microarrays in acute lymphoblastic leukemia. BMC Genomics 2004;5:71. [PubMed]
- Catchpoole D, Lail A, Guo D, et al. Gene expression profiles that segregate patients with childhood acute lymphoblastic leukaemia: an independent validation study identifies that endoglin associates with patient outcome. Leuk Res 2007;31:1741-7. [PubMed]
- Catchpoole D, Guo D, Jiang H, et al. Predicting outcome in childhood acute lymphoblastic leukemia using gene expression profiling: prognostication or protocol selection? Blood 2008;111:2486-7. [PubMed]
- Gajjar A, Hernan R, Kocak M, et al. Clinical, histopathologic, and molecular markers of prognosis: toward a new disease risk stratification system for medulloblastoma. J Clin Oncol 2004;22:984-93. [PubMed]
- Stallings RL, Carty P, McArdle L, et al. Evolution of unbalanced gain of distal chromosome 2p in neuroblastoma. Cytogenet Genome Res 2004;106:49-54. [PubMed]
- McArdle L, McDermott M, Purcell R, et al. Oligonucleotide microarray analysis of gene expression in neuroblastoma displaying loss of chromosome 11q. Carcinogenesis 2004;25:1599-609. [PubMed]
- Astuti D, Latif F, Wagner K, et al. Epigenetic alteration at the DLK1-GTL2 imprinted domain in human neoplasia: analysis of neuroblastoma, phaeochromocytoma and Wilms' tumour. Br J Cancer 2005;92:1574-80. [PubMed]
- Margetts CD, Astuti D, Gentle DC, et al. Epigenetic analysis of HIC1, CASP8, FLIP, TSP1, DCR1, DCR2, DR4, DR5, KvDMR1, H19 and preferential 11p15.5 maternal-allele loss in von Hippel-Lindau and sporadic phaeochromocytomas. Endocr Relat Cancer 2005;12:161-72. [PubMed]
- Margetts CD, Morris M, Astuti D, et al. Evaluation of a functional epigenetic approach to identify promoter region methylation in phaeochromocytoma and neuroblastoma. Endocr Relat Cancer 2008;15:777-86. [PubMed]
- Chetcuti A, Aktas S, Mackie N, et al. Expression profiling reveals MSX1 and EphB2 expression correlates with the invasion capacity of Wilms tumors. Pediatr Blood Cancer 2011;57:950-7. [PubMed]
- Natrajan R, Little SE, Sodha N, et al. Analysis by array CGH of genomic changes associated with the progression or relapse of Wilms' tumour. J Pathol 2007;211:52-9. [PubMed]
- Natrajan R, Reis-Filho JS, Little SE, et al. Blastemal expression of type I insulin-like growth factor receptor in Wilms' tumors is driven by increased copy number and correlates with relapse. Cancer Res 2006;66:11148-55. [PubMed]
- Natrajan R, Little SE, Reis-Filho JS, et al. Amplification and overexpression of CACNA1E correlates with relapse in favorable histology Wilms' tumors. Clin Cancer Res 2006;12:7284-93. [PubMed]
- Natrajan R, Warren W, Messahel B, et al. Complex patterns of chromosome 9 alterations including the p16INK4a locus in Wilms tumours. J Clin Pathol 2008;61:95-102. [PubMed]
- de Bock CE, Ardjmand A, Molloy TJ, et al. The Fat1 cadherin is overexpressed and an independent prognostic factor for survival in paired diagnosis-relapse samples of precursor B-cell acute lymphoblastic leukemia. Leukemia 2012;26:918-26. [PubMed]
- Ardjmand A, de Bock CE, Shahrokhi S, et al. Fat1 cadherin provides a novel minimal residual disease marker in acute lymphoblastic leukemia. Hematology 2013;18:315-22. [PubMed]
- Stallings RL, Nair P, Maris JM, et al. High-resolution analysis of chromosomal breakpoints and genomic instability identifies PTPRD as a candidate tumor suppressor gene in neuroblastoma. Cancer Res 2006;66:3673-80. [PubMed]
- Dunwell TL, Dickinson RE, Stankovic T, et al. Frequent epigenetic inactivation of the SLIT2 gene in chronic and acute lymphocytic leukemia. Epigenetics 2009;4:265-9. [PubMed]
- Dunwell TL, Hesson LB, Pavlova T, et al. Epigenetic analysis of childhood acute lymphoblastic leukemia. Epigenetics 2009;4:185-93. [PubMed]
- Hesson LB, Dunwell TL, Cooper WN, et al. The novel RASSF6 and RASSF10 candidate tumour suppressor genes are frequently epigenetically inactivated in childhood leukaemias. Mol Cancer 2009;8:42. [PubMed]
- Dunwell T, Hesson L, Rauch TA, et al. A genome-wide screen identifies frequently methylated genes in haematological and epithelial cancers. Mol Cancer 2010;9:44. [PubMed]
- Hill VK, Dunwell TL, Catchpoole D, et al. Frequent epigenetic inactivation of KIBRA, an upstream member of the Salvador/Warts/Hippo (SWH) tumor suppressor network, is associated with specific genetic event in B-cell acute lymphocytic leukemia. Epigenetics 2011;6:326-32. [PubMed]
- Hu M, Fletcher J, McCahon E, et al. Bilateral Wilms tumor and early presentation in pediatric patients is associated with the truncation of the Wilms tumor 1 protein. J Pediatr 2013;163:224-9. [PubMed]
- McIver SC, Stanger SJ, Santarelli DM, et al. A unique combination of male germ cell miRNAs coordinates gonocyte differentiation. PLoS One 2012;7:e35553. [PubMed]
- McIver SC, Loveland KL, Roman SD, et al. The chemokine CXCL12 and its receptor CXCR4 are implicated in human seminoma metastasis. Andrology 2013;1:517-29. [PubMed]
- Wang D, Owler BK. Expression of AQP1 and AQP4 in paediatric brain tumours. J Clin Neurosci 2011;18:122-7. [PubMed]
- Karsa M, Dalla Pozza L, Venn NC, et al. Improving the identification of high risk precursor B acute lymphoblastic leukemia patients with earlier quantification of minimal residual disease. PLoS One 2013;8:e76455. [PubMed]
- Lau LM, Dagg RA, Henson JD, et al. Detection of alternative lengthening of telomeres by telomere quantitative PCR. Nucleic Acids Res 2013;41:e34. [PubMed]
- Roma J, Masià A, Reventós J, et al. Notch pathway inhibition significantly reduces rhabdomyosarcoma invasiveness and mobility in vitro. Clin Cancer Res 2011;17:505-13. [PubMed]
- Swarbrick A, Woods SL, Shaw A, et al. miR-380-5p represses p53 to control cellular survival and is associated with poor outcome in MYCN-amplified neuroblastoma. Nat Med 2010;16:1134-40. [PubMed]
- Taylor JG 6th, Cheuk AT, Tsang PS, et al. Identification of FGFR4-activating mutations in human rhabdomyosarcomas that promote metastasis in xenotransplanted models. J Clin Invest 2009;119:3395-407. [PubMed]
- Guo X, Chen QR, Song YK, et al. Exon array analysis reveals neuroblastoma tumors have distinct alternative splicing patterns according to stage and MYCN amplification status. BMC Med Genomics 2011;4:35. [PubMed]
- Miller S, Rogers HA, Lyon P, et al. Genome-wide molecular characterization of central nervous system primitive neuroectodermal tumor and pineoblastoma. Neuro Oncol 2011;13:866-79. [PubMed]
- Picard D, Miller S, Hawkins CE, et al. Markers of survival and metastatic potential in childhood CNS primitive neuro-ectodermal brain tumours: an integrative genomic analysis. Lancet Oncol 2012;13:838-48. [PubMed]
- Henriquez NV, Forshew T, Tatevossian R, et al. Comparative expression analysis reveals lineage relationships between human and murine gliomas and a dominance of glial signatures during tumor propagation in vitro. Cancer Res 2013;73:5834-44. [PubMed]
- Rogers HA, Ward JH, Miller S, et al. The role of the WNT/β-catenin pathway in central nervous system primitive neuroectodermal tumours (CNS PNETs). Br J Cancer 2013;108:2130-41. [PubMed]
- Whittier KL, Boese EA, Gibson-Corley KN, et al. G-protein coupled receptor expression patterns delineate medulloblastoma subgroups. Acta Neuropathol Commun 2013;1:66. [PubMed]
- Brohl AS, Solomon DA, Chang W, et al. The genomic landscape of the Ewing Sarcoma family of tumors reveals recurrent STAG2 mutation. PLoS Genet 2014;10:e1004475. [PubMed]
- Shern JF, Chen L, Chmielecki J, et al. Comprehensive genomic analysis of rhabdomyosarcoma reveals a landscape of alterations affecting a common genetic axis in fusion-positive and fusion-negative tumors. Cancer Discov 2014;4:216-31. [PubMed]
- Chakravadhanula M, Hampton CN, Chodavadia P, et al. Wnt pathway in atypical teratoid rhabdoid tumors. Neuro Oncol 2015;17:526-35. [PubMed]
- Chakravadhanula M, Ozols VV, Hampton CN, et al. Expression of the HOX genes and HOTAIR in atypical teratoid rhabdoid tumors and other pediatric brain tumors. Cancer Genet 2014;207:425-8. [PubMed]
- Vaught JB, Henderson MK, Compton CC. Biospecimens and biorepositories: from afterthought to science. Cancer Epidemiol Biomarkers Prev 2012;21:253-5. [PubMed]
- Basik M, Aguilar-Mahecha A, Rousseau C, et al. Biopsies: next-generation biospecimens for tailoring therapy. Nat Rev Clin Oncol 2013;10:437-50. [PubMed]
- Tupasela A, Stephens N. The boom and bust cycle of biobanking - thinking through the life cycle of biobanks. Croat Med J 2013;54:501-3. [PubMed]