Molecular genetic studies of complete hydatidiform moles
Introduction
Complete hydatidiform moles (CHM) have no identifiable embryonic or fetal tissues and result from an abnormal growth of trophoblastic cells that would otherwise normally develop into the placenta. The name hydatidiform mole comes from the Greek words hydatisia which means drop of water and mola which means false conception. CHM often present with vaginal bleeding and ultrasound examination shows widespread and marked hyperplasia with swollen villi (1,2). They most often have the karyotype 46,XX with both haploid sets of chromosomes being paternally derived (3).
CHM fall under the categorisation of gestational trophoblastic disease, which also includes partial hydatidiform moles and gestational trophoblastic tumours (invasive mole, gestational choriocarcinoma, placental site trophoblastic tumour and epithelioid trophoblastic tumour) (4). CHM can be distinguished from partial moles by genetics (partial moles are triploid), morphology, and immunostaining of maternally expressed genes (2).
Recent epidemiological data is scarce and previously reported data on the incidence of hydatidiform moles (CHM and PHM) is variable. In North America and Europe the incidence ranges from 0.6-1.1 per 1,000 pregnancies, in Southeast Asia and Japan the incidence is as high as 2.0 per 1,000 pregnancies, and in Australia ranges from 0.91-1.41 per 1,000 pregnancies (5) with CHM at 1 in 2,000 pregnancies (6-8). The variability is primarily due to inconsistent case definitions, no centralised database, and the rarity of certain forms of molar pregnancies. Maternal age appears to be the main risk factor with women at either end of reproductive age having the highest risk. Women younger than 16 have a 6-fold increased risk and women older than 40 have a 5-10 fold increased risk of hydatidiform mole compared to women aged 16-40 years. Women aged over 50 years have a 1 in 3 risk of complete molar pregnancy (9,10). History of a previous CHM is also a risk factor and such women have a 10-20-fold higher risk than that of the general population (11).
The genetic basis of hydatidiform moles was established in the 1970’s (3). All CHM have a diandric paternal only genome; there is no maternal contribution. The majority of CHM are therefore homozygous and arise from an anuclear empty ovum that has been fertilised by a haploid 23,X sperm (monospermy), which then replicates its own chromosomes resulting in a 46,XX karyotype (uniparental paternal isodisomy). A minority involve the fertilisation of an anuclear empty ovum with two sperm (dispermy) simultaneously, which can result in a 46,XX or 46,XY karyotype (uniparental paternal heterodisomy) (12). A very rare form of CHM involves tetraploidy (13). It has also been reported that diploidization may follow from a triploid conceptus; a CHM would result if the maternal haploid contribution was lost (14). The abnormality in fetal-placental development in CHM is essentially due to abnormal genomic imprinting effects. Loss of the maternal epigenetic imprint and gain of paternally imprinted gene expression together result in global genome demethylation and abnormal gene expression that results in abnormal placental trophoblast development (15).
Most cases of CHM are sporadic, however a very small minority of moles are recurrent and often familial, usually being diploid and biparental (having both sets of parental chromosomes). Rare recurrent hydatidiform moles (RHM) involve at least two molar pregnancies. Maternal homozygous and compound heterozygous recessive gene mutations have been reported in some families. Two such genes are NLRP7 and KHDC3L (16). NLRP7 (NLR family, pyrin domain containing 7) located at 19q13.4 is a member of the NLR family of proteins with a role in inflammation and apoptosis (17). It was the first identified recessive gene involved in RHMs and mutations have been reported in 48-80% of patients with more than 50 mutations identified to date (18-20). KHDC3L (KH domain containing 3-like) located at 6q13 is a member of the KHDC1 protein family (21). It was the second recessive gene identified responsible for RHM, however only five mutations have been identified to date (22). The exact causal mechanisms involving these two genes in RHM families is not fully understood, however deregulation of imprinted genes contributes to pathogenesis, resulting in aberrant cell proliferation and differentiation (16).
The clinical features of CHM in the first trimester are less obvious than in the second trimester with less vaginal bleeding, and excessive uterine size only in a minority of patients. Ultrasound shows less cavitation and smaller villi. Interestingly, the diagnosis of CHM has moved from second trimester to first trimester, often before classic signs and symptoms present. This is due to more accurate and sensitive detection of β-hCG and improved early ultrasonographic examination (3). If molar characteristics are not present at early ultrasound, then a missed abortion may be suspected rather than a molar pregnancy, although a strikingly elevated β-hCG can help recognize a CHM due to the hyperplastic trophoblastic cells of a molar pregnancy (23).
Conventional cytogenetics, once considered the gold standard of genetic investigation cannot distinguish a 46,XX or 46,XY CHM from a 46,XX or 46,XY non-molar conceptus. Accurate diagnosis of CHM is of clinical importance because of the increased risk (18-29%) of developing gestational trophoblastic neoplasia. On the other-hand, wrong classification of a CHM can lead to unnecessary surveillance and delay in attempting another pregnancy. The move to early diagnosis means that clinical diagnosis alone can wrongly classify CHM as partial moles or non-molar abortions (3). Diagnosis can be improved by including molecular genotyping using either polymorphic STR analysis, or alternatively analysis by whole genome SNP microarray. Although this strategy can also detect triploid ‘partial moles’, this is not the focus of the discussion within this paper. We report the results from 3 years of products of conceptions (POC) testing with the aim to refine our molecular genetic testing strategy for the identification of CHM among POCs referred for a wide range of indications.
Materials and methods
POC specimens
Referrals for genetic testing of POC and stillbirths were received from both public hospitals and private clinics from around New South Wales, Australia. Tissues received were examined macroscopically and dissected using a bifocal dissection microscope. Identifiable fetal tissues were preferentially dissected and selected for testing, however when not available, other non-maternal tissues were selected including chorionic villus (CV) and fetal membranes that were cleaned of blood clots and maternal decidua. Between January 2012 and December 2014, a total of 713 specimens were referred for genetic investigation, however following tissue dissection, only 643 specimens proceeded to testing. Gestations ranged from 7-41 weeks. Any specimens with clinical suspicion of ‘complete’ or ‘partial’ hydatidiform mole and/or ‘hydropic’ chorionic villus [placenta] identified at the time of dissection were noted. These specimens (n=32) subsequently underwent STR genotyping analysis.
DNA extraction
Tissue was macerated by mincing with a scalpel, digested at 37 °C with Protease (Qiagen), and DNA extraction performed using a modified salting out protocol (24). Resulting DNA was then quantified using a Nanodrop spectrophotometer to determine concentration and quality values. DNA that was of insufficient quality as determined by 260/280 and 260/230 ratios underwent additional purification steps involving RNAse (Qiagen) and Protease (Qiagen) treatments followed by re-precipitation.
Microarray comparative genomic hybridisation (CGH)
All samples were analysed by genome-wide microarray CGH using Agilent’s SurePrint G3 ‘ISCA Targeted’ Microarray (8×60K), according to the manufacturer’s protocol and using a commercial male control DNA (Promega) but with slight modification. Following Rsa1/Alu1 digestion, fragmented DNA underwent labelling by random priming using Klenow and either dUTP-Cy5 (sample) or dUTP-Cy3 (reference) in a 200 µL reaction incubated overnight at 37 °C, according to our standard laboratory protocol. Microarray slides were scanned using Agilent’s Feature Extraction software v10.7.1.1 and quality metrics of microarray hybridisation data were assessed prior to further analysis. Agilent’s Genomic Workbench Standard Edition Version 5.0.14, or Cytogenomics v2.7.1.0 was then used to analyse the microarray data at an effective mean resolution of 1.0 Mb. Therefore our reporting threshold for chromosome copy number change was ≥1.0 Mb in size.
Genotyping
Genotyping of microsatellite short tandem repeat (STR) markers was performed by Quantitative Fluorescent Polymerase Chain Reaction (QF-PCR) amplification using two different commercially available approaches, either using QST*R v2plus (Elucigene) or PowerPlex 16 HS (Promega), according to the manufacturers protocols. PCR products were then separated using capillary electrophoresis (Applied BioSystems), allowing for allele sizing and dosage to be performed using GeneMarker (Softgenetics). Complete molar pregnancies were defined when single (homozygous) alleles were evident across all autosomal STR markers. Partial molar pregnancy or triploidy was defined when informative autosomal STR markers indicated triallelic patterns as either 1:2, 2:1 or 1:1:1 relative peak area ratios. Although both kits can be used interchangeably to detect molar pregnancies and maternal cell contamination, we preferentially used the QST*R kit when a molar pregnancy was suspected, either from clinical notes or when ‘hydropic’ villi are identified, and the PowerPlex 16 HS kit to exclude maternal cell contamination for other placental tissue specimens.
Results
Overall, 643 POC were analysed by microarray CGH which included 272 first-trimester, 258 second-trimester and 113 third-trimester referrals. A total of 32 samples suspected of being molar underwent supplementary genotyping by STR analysis, with 10/32 (31.2%) identified among suspected samples giving an incidence of 3.6% (10/272) amongst first trimester specimens. Interestingly, although other chromosomal abnormalities occurred more frequently, CHM were the ninth most frequent chromosomal anomaly and were detected in 10/643 (1.5%) specimens and were slightly more frequent than partial ‘triploid’ moles, which were found in 7/643 (1.1%) specimens.
Table 1 shows the sample numbers and abnormality rates broken down into first-, second- and third trimester referrals. In Figure 1, the distribution of the different chromosomal abnormalities among all POC specimens can be seen with trisomy 21 being the most frequent followed by other abnormalities in decreasing frequency.
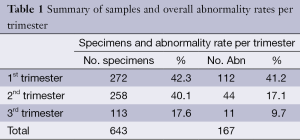
Full table
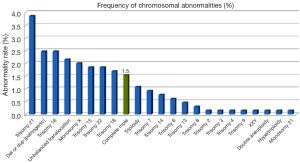
Discussion
Chromosomal and genetic anomalies have a significant adverse effect on human reproduction, commonly resulting in miscarriage and stillbirths and genetic investigation can provide important information on reproductive health risks. A number of techniques have been used to characterise the genetic status of POCs. Chromosome analysis (karyotyping) examines all 46 chromosomes and has been considered the principal investigation for causes of pregnancy loss (25) for several decades [since the 1970s]. Fluorescent in situ hybridisation (FISH) uses fluorescently tagged probes to visualise specific DNA segments and has also been applied more recently, however only a limited number of chromosomes (five) are typically examined (26,27). Later molecular techniques that can identify chromosome aneuploidy have included multiplex ligation-dependent probe amplification (MLPA) (28), microarray CGH (29), or whole genome SNP microarray. DNA ploidy analysis can also be performed by flow cytometry (30). In contrast to MLPA and microarray CGH, triploidy (3n) and diploidy (2n) moles can be distinguished by karyotyping, FISH and flow cytometry, however none of these techniques can determine parental origin of the chromosomes and distinguish between a CHM and a non-molar abortus. Alternatively, genotyping using STR microsatellite polymorphisms, or whole genome SNP microarray, can determine chromosome parental origin.
STRs are repetitive polymorphic DNA sequences, prevalent in non-coding regions. They are genetically stable and the number of polymorphic repeats differs between individuals and therefore can be used to create a genetic profile or genotype. This is helpful to identify the parental origin of chromosomes in molar pregnancies. The majority of complete 46,XX moles have complete uniparental isodisomy for all chromosomes. They will have only one homozygous allele size at each loci (Figure 2); thereby allowing them to be distinguished from non-molar (heterozygous) 46,N conceptuses, which would be expected to show two alleles at a number of loci; although some loci may be non-informative where both alleles show the same repeat size. Complete moles resulting from dispermy will be biallelic with both sets of chromosomes paternal (heterodisomy). However, these will not be distinguished from non-molar [heterozygous] pregnancies without comparing to a maternal or paternal genotype profile (13,31).
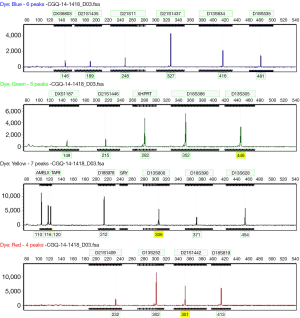
Our genomes contain single nucleotide polymorphisms (SNPs) and SNP arrays were developed to allow genotyping of thousands of SNPs across the genome simultaneously. They can also detect genomic copy number variation such as submicroscopic deletion and duplication, amplification, absence of heterozygosity (AOH), and uniparental disomy (32). SNP microarray analysis that detects AOH for all chromosomes identifies uniparental isodisomy consistent with a CHM. For cases of CHM resulting from dispermy, as noted above, comparison to maternal or paternal SNP genotype data is needed to exclude a maternal contribution.
We investigated the incidence of CHM from a cross-sectional survey on 643 miscarriage and stillbirth specimens from all trimesters of pregnancy. A range of chromosomal abnormalities were detected and the frequency of abnormalities decreased from 41.2% in the first-trimester to 9.7% in the third-trimester. We note that during the study period we did not apply STR genotype analysis to all placental samples, only to those with suspicion of a molar pregnancy (n=32). Therefore, the number of molar pregnancies may be underestimated and we cannot confidently exclude maternal cell contamination among other placental specimens, however we note that the overall normal male to female sex ratio in our data was 0.9 (231 male:244 female).
Abnormalities involving whole or segmental chromosome copy number change were easily detected by microarray CGH, however the ‘complete moles’ and partial triploidy moles gave normal microarray CGH results, since microarray CGH cannot detect changes of chromosome ploidy; e.g., diploidy, triploidy or tetraploidy. The molar pregnancies observed in our study were detected following STR genotype analysis, which was performed because of either clinical suspicion or the presence of ‘hydropic’ villus. Of the ten complete moles identified, nine were suspected clinically following ultrasound findings and were accompanied by clinical notes indicating ‘complete mole’ or ‘molar pregnancy’. One other had clinical notes indicating ‘absent yolk sac/fetal parts’ but showed ‘hydropic villi’ at tissue dissection. All of these 10 specimens were identified at an early gestation <10 weeks. Interestingly, of the 7 partial ‘triploid’ moles identified, 2 had clinical notes of early miscarriage <9 weeks, one was described as ‘partial mole’, one was a termination following ‘abnormal ultrasound’ findings within the second trimester, and the remainder were described as ‘miscarriage’. Although only one referral suggested a ‘partial mole’, the 7 specimens showed hydropic villus at tissue dissection and therefore subsequently underwent STR analysis.
Since 2010, we replaced karyotyping with microarray CGH for investigation of miscarriage and stillbirths, which saw improvements in test success and detection rates and eliminated labour intensive tissue culture and chromosome analysis by microscopy (unpublished data). Our laboratory transitioned prenatal rapid aneuploidy screening from a FISH to QF-PCR STR based assay from January to June 2012, which also allowed analysis of POC specimens. Altogether, our data shows that STR genotyping has good diagnostic utility when a complete mole is suspected clinically and when hydropic villi are found at tissue dissection. However, any molar pregnancies without a clinical suspicion would still go undiagnosed with this approach. Therefore, we have implemented a revised testing strategy used on all POC and stillbirth samples, irrespective of the clinical indication. Our testing strategy is outlined in Figure 3.
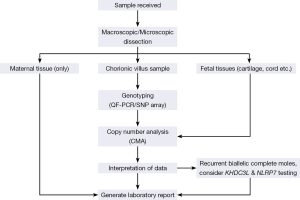
POCs are received by the laboratory and cleaned and dissected free of blood clots and maternal decidua. No further testing proceeds if only maternal tissue is identified. A laboratory report is subsequently issued accordingly. When identifiable fetal parts (e.g., skin, cord or cartilage etc.) are identified, testing proceeds directly to microarray CGH genome wide analysis. When no fetal parts are identified and there is only placental chorionic villus material available, we reflex to STR genotyping prior to microarray analysis to identify maternal cell contamination, partial hydatidiform moles and AOH consistent with CHM (any aneuploidies detected by STR analysis will proceed to microarray CGH for confirmation). We note that if genotyping was performed using whole genome SNP array analysis then targeted STR analysis would not be required.
This testing strategy has the limitation that if a 46,XX or 46,XY biallelic result was obtained and a molar pregnancy was still strongly suspected, genotype ‘trio’ analysis with both parental (maternal and paternal) profiles may be necessary for diagnosis; however we note parental samples are rarely forthcoming amongst miscarriage referrals. For the rare cases of biparental complete moles, diagnosis is usually made on a clinical basis, however other ancillary testings such as p57 immunochemistry (33) and specific molecular testing may be considered for recurrent moles; e.g., mutations of NLRP7 and KHDC3L. Unfortunately, we currently do not offer testing for these genes.
Apart from identifying molar pregnancies, the benefits of molecular testing by microarray CGH supplemented with QF-PCR STR genotyping [or solely by SNP genotyping microarray] include the use of DNA extracted directly from uncultured samples, thereby avoiding culture bias and failure, which can further result in improved turnaround times and increased abnormality detection rates, as well as provide a means to exclude maternal cell contamination.
Conclusions
Conventional cytogenetics has been the principal investigation tool of pregnancy failure since the 1970s, which can detect a wide range of chromosomal abnormalities, including triploidy partial molar pregnancies, however it cannot distinguish between CHM and non-molar pregnancies. To detect molar pregnancies, molecular cytogenomic approaches should be used including genotyping by STR or SNP microarray analysis. Furthermore, the diagnostic utility of genotyping for accurate classification of hydatidiform mole has also been recently reported (34) and genotyping can aid in clinical management and genetic counselling. A CHM is a rare finding, however it can develop into gestational trophoblastic neoplasia, a malignant and life threatening disease. Therefore the use of molecular genotyping is important and if used for all POC referrals will allow for identification of the vast majority of CHM. We further suggest that for rare recurrent (2 or more) biparental complete moles that mutation testing of NLRP7 and KHDC3L should be considered.
Acknowledgements
The authors would like to thank the Nucleic Acid Unit for DNA extractions and the Prenatal and Microarray laboratories for sample processing within Sydney Genome Diagnostics, Western Sydney Genetics Program at The Children’s Hospital at Westmead.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Monchek R, Wiedaseck S. Gestational trophoblastic disease: an overview. J Midwifery Womens Health 2012;57:255-9. [PubMed]
- Berkowitz RS, Goldstein DP. Clinical practice. Molar pregnancy. N Engl J Med 2009;360:1639-45. [PubMed]
- Kajii T, Ohama K. Androgenetic origin of hydatidiform mole. Nature 1977;268:633-4. [PubMed]
- Hoffner L, Surti U. The genetics of gestational trophoblastic disease: a rare complication of pregnancy. Cancer Genet 2012;205:63-77. [PubMed]
- Bracken MB. Incidence and aetiology of hydatidiform mole: an epidemiological review. Br J Obstet Gynaecol 1987;94:1123-35. [PubMed]
- Jeffers MD, O'Dwyer P, Curran B, et al. Partial hydatidiform mole: a common but underdiagnosed condition. A 3-year retrospective clinicopathological and DNA flow cytometric analysis. Int J Gynecol Pathol 1993;12:315-23. [PubMed]
- Palmer JR. Advances in the epidemiology of gestational trophoblastic disease. J Reprod Med 1994;39:155-62. [PubMed]
- Khoo SK. Clinical aspects of gestational trophoblastic disease: a review based partly on 25-year experience of a statewide registry. Aust N Z J Obstet Gynaecol 2003;43:280-9. [PubMed]
- Sebire NJ, Seckl MJ. Gestational trophoblastic disease: current management of hydatidiform mole. BMJ 2008;337:a1193. [PubMed]
- Bagshawe KD, Lawler SD. Unmasking moles. Br J Obstet Gynaecol 1982;89:255-7. [PubMed]
- Tse KY, Chan KK, Tam KF, et al. 20-year experience of managing profuse bleeding in gestational trophoblastic disease. J Reprod Med 2007;52:397-401. [PubMed]
- Lawler SD, Fisher RA. Dent J. A prospective genetic study of complete and partial hydatidiform moles. Am J Obstet Gynecol 1991;164:1270-7. [PubMed]
- Hui P. Molecular diagnosis of gestational trophoblastic disease. Expert Rev Mol Diagn 2010;10:1023-34. [PubMed]
- Golubovsky MD. Postzygotic diploidization of triploids as a source of unusual cases of mosaicism, chimerism and twinning. Hum Reprod 2003;18:236-42. [PubMed]
- Bestor TH, Bourc'his D. Genetics and epigenetics of hydatidiform moles. Nat Genet 2006;38:274-6. [PubMed]
- Nguyen NM, Slim R. Genetics and Epigenetics of Recurrent Hydatidiform Moles: Basic Science and Genetic Counselling. Curr Obstet Gynecol Rep 2014;3:55-64. [PubMed]
- Murdoch S, Djuric U, Mazhar B, et al. Mutations in NALP7 cause recurrent hydatidiform moles and reproductive wastage in humans. Nat Genet 2006;38:300-2. [PubMed]
- Slim R, Bagga R, Chebaro W, et al. A strong founder effect for two NLRP7 mutations in the Indian population: an intriguing observation. Clin Genet 2009;76:292-5. [PubMed]
- Hayward BE, De Vos M, Talati N, et al. Genetic and epigenetic analysis of recurrent hydatidiform mole. Hum Mutat 2009;30:E629-39. [PubMed]
- Estrada H, Buentello B, Zenteno JC, et al. The p.L750V mutation in the NLRP7 gene is frequent in Mexican patients with recurrent molar pregnancies and is not associated with recurrent pregnancy loss. Prenat Diagn 2013;33:205-8. [PubMed]
- Parry DA, Logan CV, Hayward BE, et al. Mutations causing familial biparental hydatidiform mole implicate c6orf221 as a possible regulator of genomic imprinting in the human oocyte. Am J Hum Genet 2011;89:451-8. [PubMed]
- Reddy R, Akoury E, Phuong Nguyen NM, et al. Report of four new patients with protein-truncating mutations in C6orf221/KHDC3L and colocalization with NLRP7. Eur J Hum Genet 2013;21:957-64. [PubMed]
- Sebire NJ, Fisher RA, Foskett M, et al. Risk of recurrent hydatidiform mole and subsequent pregnancy outcome following complete or partial hydatidiform molar pregnancy. BJOG 2003;110:22-6. [PubMed]
- Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1988;16:1215. [PubMed]
- Lomax B, Tang S, Separovic E, et al. Comparative genomic hybridization in combination with flow cytometry improves results of cytogenetic analysis of spontaneous abortions. Am J Hum Genet 2000;66:1516-21. [PubMed]
- Yver M, Carles D, Bloch B, et al. Determination of DNA ploidy by fluorescence in situ hybridization (FISH) in hydatidiform moles: evaluation of FISH on isolated nuclei. Hum Pathol 2004;35:752-8. [PubMed]
- Lescoat D, Jouan H, Loeuillet-Olivo L, et al. Fluorescent in situ hybridization (FISH) on paraffin-embedded placental tissues as an adjunct for understanding the etiology of early spontaneous abortion. Prenat Diagn 2005;25:314-7. [PubMed]
- Caramins MC, Saville T, Shakeshaft R, et al. A comparison of molecular and cytogenetic techniques for the diagnosis of pregnancy loss. Genet Med 2011;13:46-51. [PubMed]
- Schaeffer AJ, Chung J, Heretis K, et al. Comparative genomic hybridization-array analysis enhances the detection of aneuploidies and submicroscopic imbalances in spontaneous miscarriages. Am J Hum Genet 2004;74:1168-74. [PubMed]
- Lage JM, Popek EJ. The role of DNA flow cytometry in evaluation of partial and complete hydatidiform moles and hydropic abortions. Semin Diagn Pathol 1993;10:267-74. [PubMed]
- Murphy KM, McConnell TG, Hafez MJ, et al. Molecular genotyping of hydatidiform moles: analytic validation of a multiplex short tandem repeat assay. J Mol Diagn 2009;11:598-605. [PubMed]
- LaFramboise T. Single nucleotide polymorphism arrays: a decade of biological, computational and technological advances. Nucleic Acids Res 2009;37:4181-93. [PubMed]
- Castrillon DH, Sun D, Weremowicz S, et al. Discrimination of complete hydatidiform mole from its mimics by immunohistochemistry of the paternally imprinted gene product p57KIP2. Am J Surg Pathol 2001;25:1225-30. [PubMed]
- Furtado LV, Paxton CN, Jama MA, et al. Diagnostic utility of microsatellite genotyping for molar pregnancy testing. Arch Pathol Lab Med 2013;137:55-63. [PubMed]


