
ABCA3 gene mutations shape the clinical profiles of severe unexplained respiratory distress syndrome in late preterm and term infants
Introduction
Neonatal respiratory distress syndrome (RDS) is caused by the deficiency or inactivation of pulmonary surfactant, and is commonly seen in early preterm infants due to their immature lung development. However, some maternal and neonatal characteristics in late preterm and term infants such as maternal diabetes, meconium aspirated pneumonia, neonatal sepsis, and severe intrapartum asphyxia, could also contribute to RDS. Existing evidence has demonstrated that RDS in late preterm and term infants is a somewhat distinct disease entity, with risk factors and clinical profiles that differ from those in early preterm infants (1-3). In a neonatal unit, some late preterm or term infants experience RDS; however, the etiology has not been well defined after a routine workup. This unique RDS entity was referred to as unexplained respiratory distress syndrome (URDS) by numerous previous studies, as well as our study (4-7).
Given that a large body of literature has demonstrated the essential role genetic mechanism play in the pathogenesis of most URDS cases, numerous medical facilities have carried out clinical exome sequencing to identify the underlying genetic cause of URDS (8-10). Among all of the genetic factors that contribute to the RDS, the most common is ABCA3 gene mutation, which involves the assembly of pulmonary surfactant in the lamellar body of pneumocyte II (11). Existing evidence indicated that patients with homozygous or compound heterozygous ABCA3 gene mutations were commonly in critically ill conditions (12). A single ABCA3 mutation was also likely to increase the risk and severity of RDS (4,13).
Unfortunately, in a considerable proportion of neonatal URDS patients, genetic testing fails to yield any abnormal findings, making these patients the “true URDS” patients. Currently, whether the ABCA3-mutated URDS patients have similar or more challenging clinical profiles to those without any genetic abnormalities continues to confound most neonatologists. An answer to this question would help to guide the management and predict the clinical outcomes of neonatal URDS patients. The present study aimed to address this by comparing the clinical characteristics of late preterm and term infants with severe URDS with homozygous or compound heterozygous ABCA3 mutations, a single ABCA3 mutation, or no defined genetic abnormalities.
We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/tp-20-283).
Methods
Patient selection
This single-center retrospective cohort study involved infants ≥34 weeks’ gestation with severe URDS who were admitted to Children’s Hospital of Chongqing Medical University between January 2013 and December 2019. In this study, severe RDS was mainly defined according to the consensus of the Pediatric Acute Lung Injury Consensus Conference (14), and Montreux definition (15): (I) manifestations and chest radiograph compatible with RDS; (II) on invasive mechanical ventilation with oxygenation index ≥16, which was calculated based on the daily blood gas, or as an alternative measurement, on the subcutaneous oxygen tension (16,17).
Almost all infants with severe RDS had undergone a comprehensive workup, including serial infection markers, chest radiograph, echocardiography, and blood and sputum pathogen testing. For all patients with severe RDS who responded inadequately to interventions and had unremarkable workup findings, trio exome sequencing on samples from patients and their parents were usually recommended. All URDS patients who underwent genetic testing were enrolled in this study. Those whose parents rejected genetic testing, or who had cardiopulmonary malformations, pulmonary hypoplasia, culture-positive sepsis, or known respiratory disease-associated gene mutations (such as SFTPA1, SFTPA2, SFTPB, SFTPC, CHPT1, LPCAT1, PCYT1B, NKX2, CFTR, and FOXF1) were excluded. This study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Institutional Review Board of Children’s Hospital of the Chongqing Medical University (No. 2018-158) and was registered on clinicaltrials.gov (NCT04137783).
Exome sequencing and gene mutation detection
All infants in this study underwent trio exome sequencing after written consent had been obtained from their parents. A gene company (Chigene, Beijing) offered the sequencing as a clinical laboratory service. Sequence analysis of coding exons and flanking introns were performed as previously described (18,19). All samples were analyzed to detect frame-shift mutations, nonsense mutations, missense mutations, splicing site mutations, and in-frame indel mutations. Assessment of copy number variation was also performed from exome sequencing data using computational tools. A variant was strictly defined as a mutation if it had been previously described to cause disease with a presentation consistent with these patients, or resulted in an amino acid change or protein structure alteration to disrupt protein function that was predicted by both SIFT and PolyPhen for missense mutations (20,21), and MaxEntScan and dbscSNV for splicing site mutations (22). In the case of a novel mutation, phastCons and phyloP were used to determine the evolutionary conservation of the region where the mutation was located (23). The American College of Medical Genetics and Genomics/Association for Molecular Pathology (ACMG-AMP) criteria were applied to interpret the mutations (24). The subjects in this study were categorized into three groups: homozygous or compound heterozygous ABCA3 mutations, a single ABCA3 mutation, and no defined genetic abnormalities.
Clinical profiles data
All relevant clinical data were extracted from a hospital information system. All antepartum and postpartum data were collected and included: maternal age, parity, and pregnancy-related complications; mode of birth, amniotic fluid condition, and history of asphyxia and resuscitation; postnatal age of respiratory symptom onset, modalities of respiratory support, and the daily record of blood gas and subcutaneous oxygen tension; laboratory data including complete blood count, C-reactive protein, procalcitonin, blood and sputum culture, respiratory viral detection test, and genetic testing; radiographic examination including chest X-ray and echocardiogram; medications taken during the hospital stay.
Radiological scoring
All chest X-rays were reviewed on a hospital information system by one radiologist. The most severe images were scored according to the Fleischner Society criteria (25). The chest X-ray was rated in three sections on both sides of the lung: apex to the carina, carina to the lower pulmonary vein, and lower pulmonary vein to the diaphragm. The incidence of radiological features, including ground-glass opacity, reticular pattern, air bronchogram, atelectasis, and air leak, were evaluated for each lung section. Each finding was scored as 0 = none, 1 = discrete, 2=diffuse, or 3 = strong at each section. An overall cumulative score was calculated by adding the individual section scores together, with a maximum score of 18 for each patient.
Statistical analysis
Analysis of variance (ANOVA) and the Chi-square or Fisher’s exact tests were used to compare clinical characteristics and radiographic scores between different groups of patients. Tests for the differences in the age of symptom onset and the age of development of severe RDS between the groups were carried out using the log-rank test. A two-tailed P value of <0.05 was considered statistically significant.
Results
A total of 59 infants (≥34 weeks gestation) satisfied the criteria for URDS. Of these infants, 43 underwent clinical exome sequencing between 8 to 25 days after birth. Two patients with SFTPB mutations, one with SFTPC mutations, and one with cardiopulmonary malformation were excluded from the study. Finally, 39 infants were included for analysis (Figure 1). No significant difference in the gestational age, birth weight, sex, multiplets, or maternal complications were noted between the groups (Table 1).
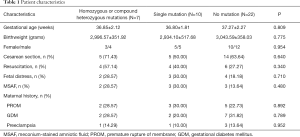
Full table
Genetic characteristics
By looking at the variants on the three patients with homozygous ABCA3 mutations, one novel intronic mutation (c.3862+1G>C) was identified in twins, which was predicted to affect the splicing site at exon 42. Both phastCons and phyloP computation indicated that the located region was highly evolutionarily conserved, with 0.954 and 0.852, respectively. Hazard predictions of the novel splicing site by MaxEntScan and dbscSNV were deleterious, with 6.97–>−1.31 and 0.9999∣0.9020, respectively. Sequencing of asymptomatic parents revealed the same variant in each of them with heterozygous expression. This was classified as pathologic according to the ACMG-AMP criteria (24). One nonsense mutation (R1561X), previously reported by Kröner et al., was identified in the third patient. Their study revealed the pathogenic characteristics of this mutation based on clinical and histological investigations (26).
For patients with compound heterozygous mutations and a single mutation, 12 missense mutations, 1 intronic mutation (c.1897-1G>C), and 1 synonymous mutation (S1372=) were identified. Interestingly, patient no. 4 and no. 5 shared a genotype (P193S/G1412R) but experienced a different clinical course; patient no. 4 died on his 24th day, whereas patient no. 5 recovered and was discharged 20 days after birth. Patient no. 16 and no. 17 also had the same genotype (E292V/−); however, they exhibited different phenotypes (Table 2).
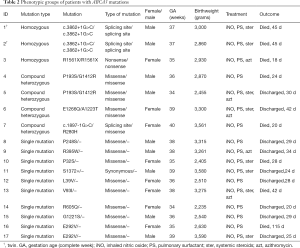
Full table
Clinical course
Of the seven patients with homozygous or compound heterozygous ABCA3 mutations, three presented with respiratory symptoms immediately after birth, while the remaining four patients presented with respiratory symptoms between 1 and 5 hours after birth, which was much earlier than those with a single ABCA3 mutation and those without genetic abnormalities (χ2=13.500, P<0.001; χ2=30.400, P<0.001). Meanwhile, all patients with homozygous or compound heterozygous mutations developed severe RDS (with an oxygenation index of 16 as an indicator) on the first day of life, which was also earlier than those with a single mutation, or a wild-type gene (χ2=6.067, P=0.014; χ2=25.150, P<0.001). However, no differences were noted between patients with a single ABCA3 mutation and those without genetic abnormalities in terms of the time of symptom onset or progression to severe RDS (χ2=1.407, P=0.236; χ2=0.304, P=0.581). All patients with URDS were given surfactant administration and nitric oxide inhalation, and some were also administered intravenous steroids and azithromycin. The subjects responded differently to these interventions (Table 2). All but two patients with homozygous or compound heterozygous mutations died at an early stage of life, and the mortality rate was marginally higher than that of infants with a single mutation, although the difference was not statistically significant (χ2=2.837, P=0.092). However, this mortality rate was markedly higher than that of infants without genetic abnormalities, five of whom died (χ2=5.575, P=0.018).
Radiological score
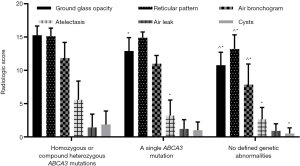
Patients with homozygous or compound heterozygous ABCA3 mutations had a higher radiological score than those with a single mutation (51.14±4.91 vs. 44.20±6.54, P=0.025). Meanwhile, patients with a single mutation had a higher radiological score than those without genetic abnormalities (44.20±6.54 vs. 35.95±4.31, P<0.001). The primary abnormalities observed in the chest X-rays in each group were ground-glass opacity, reticular pattern, and air bronchogram. Furthermore, except for air leaks, patients with homozygous or compound heterozygous mutations had significantly higher individual subset scores than those without genetic abnormalities, and their scores for ground-glass opacity and atelectasis were also higher than that of patients with a single mutation. Patients with a single mutation had higher scores for ground-glass opacity, reticular pattern, and air bronchogram than those with wild-type ABCA3 gene (Figure 2).
Discussion
Numerous studies have suggested that the ABCA3 mutation is the most common etiology of genetic RDS. A study by Brasch et al. involving 14 term infants with severe URDS (from six consanguineous families and two nonconsanguineous families) reported that 10 (71.43%) of them exhibited homozygous or compound heterozygous ABCA3 mutations (5). Similarly, Shulenin et al. reported that 16 of 21 (76.19%) infants with severe URDS from 14 families had homozygous ABCA3 gene mutations (27). Our study detected homozygous or compound heterozygous ABCA3 mutations in 4 out of 43 (9.30%) patients with severe URDS; a lower detection rate than the aforementioned studies. This discrepancy may be explained by the fact that a higher proportion of the subjects in these previous studies were twins, some with consanguineous histories.
To date, about 200 ABCA3 mutations have been identified, with the majority of them resulting in varying degrees of surfactant function impairment (28). To our knowledge, the novel splicing site (c.3862+1G>C) in our study has not been reported in any previous literature and is not documented on dbSNP or the China National Genebank Database (http://db.cngb.org). Based on the ACMG-AMP interpretation, evolutionary conservation of the affected region, and patients’ clinical phenotype, this novel splicing site was considered the etiology of RDS in twins. Most ABCA3 mutations in this study cohort were missense mutations, which could be classified into three groups based on the pathway through which they affect surfactant homeostasis. Essentially, type 1 and type 2 refer to the abnormal intracellular location and decreased adenosine triphosphate hydrolysis, respectively, while type 3 combines both type 1 and type 2 (9,28). Therefore, the diversity of clinical profiles might be attributable to the variable mutation group. Interestingly, both patients no. 4 and no. 5 had an identical mutation (P193S/G1412R) but showed different clinical characteristics. Hallik et al. reported a similar case in which two siblings expressed an identical compound heterozygous mutation but developed extremely different clinical courses of respiratory disease (29). It is thought that epigenetics, some accompanying neonatal disorders, and environmental factors play a part in the pathogenesis and prognosis of RDS to some extent.
Homozygous or compound heterozygous mutations in the ABCA3 gene have been confirmed to disrupt surfactant metabolism and have been associated with severe RDS (5,27,30). Our study found that five of the seven patients with homozygous or compound heterozygous ABCA3 mutations died at an early stage of life, which further confirmed the lethality of such mutations (5,27). Concerning clinical profiles, these patients had symptom onset and developed severe RDS much earlier than those with a single mutation or those without defined genetic abnormalities. Their radiographs were also markedly more severe. Ultrastructural analysis of neonatal lung specimens with ABCA3 homozygous or compound heterozygous mutations revealed a small lamellar body with densely packed phospholipid membranes and eccentrically placed, dense inclusion bodies (27), which is strongly indicative of immature lamellar bodies, and is predictive of surfactant impairment. Thus, the homozygous or compound heterozygous ABCA3 mutation could be strongly predictive of a challenging clinical course.
Pathological ABCA3 mutations are inherited in an autosomal recessive manner. However, numerous studies have noted that a single ABCA3 mutation could also worsen the severity of lung disease (4,13,31). Our study did not sufficiently distinguish patients with a single ABCA3 mutation from those without genetic abnormalities based on the clinical course. However, in this study, radiographic findings seemed adequate to discriminate between them. Some previous studies reported that although the ABCA3 protein could be detected in patients with a single ABCA3 mutation, the presence of type II pneumocytes hyperplasia, inflammatory cell infiltration, and lamellar bodies with smaller vesicles still suggested a deleterious effect of a single mutation on surfactant metabolism (31,32). However, the underlying mechanism of this has not been well defined. Some gene mutations might be overlooked in general clinical sequencing (33); a single ABCA3 mutation might couple with other gene mutations on the opposite allele to affect surfactant homeostasis (34).
The limitations of our study are primarily attributable to the characteristics of a retrospective study. We did not enroll all patients with URDS in our clinical setting. It is uncertain to what extent the genetic and clinical data of the missed 16 patients would have altered the current results. Also, we did not perform a long-term follow-up study. Some literature has revealed that a single ABCA3 mutation is correlated with a higher risk of interstitial lung disease at a late stage (31,33). Thus, a long-term follow-up study might identify more unique characteristics of patients with a single ABCA3 mutation compared to those without genetic abnormalities. Meanwhile, our study only focused on the ABCA3 gene, and did not address all respiratory disease-associated genetic abnormalities. However, most respiratory-associated genes have not been associated with severe RDS, and in a clinical setting, the ABCA3 gene mutation contributes to the majority of severe genetic RDS cases.
In conclusion, our study suggests that in a neonatal clinical setting of caring for severe URDS patients, identification of homozygous or compound heterozygous ABCA3 gene mutations commonly predicted more challenging clinical profiles and poor outcomes compared to patients without genetic abnormalities. Early aggressive treatment or innovative interventions should be applied to treat these patients. However, whether the same relationship exists between patients with a single ABCA3 mutation and those with no defined genetic abnormalities warrants further study.
Acknowledgments
The language in this paper has been re-edited and polished by AME editing service.
Funding: This work was supported by the Chongqing Science and Technology Commission (No. cstc2018jscx-msybX0071).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/tp-20-283
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tp-20-283
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tp-20-283). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Institutional Review Board of Children’s Hospital of Chongqing Medical University (No. 2018-158) and individual consent for this retrospective analysis was waived. All infants in this study underwent trio exome sequencing after written consent had been obtained from their parents.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Wang J, Liu X, Zhu T, et al. Analysis of neonatal respiratory distress syndrome among different gestational segments. Int J Clin Exp Med 2015;8:16273-9. [PubMed]
- Shi Y, Tang S, Zhao J, et al. A prospective, randomized, controlled study of NIPPV versus nCPAP in preterm and term infants with respiratory distress syndrome. Pediatr Pulmonol 2014;49:673-8. [Crossref] [PubMed]
- Condò V, Cipriani S, Colnaghi M, et al. Neonatal respiratory distress syndrome: are risk factors the same in preterm and term infants? J Matern Fetal Neonatal Med 2017;30:1267-72. [Crossref] [PubMed]
- Somaschini M, Presi S, Ferrari M, et al. Surfactant proteins gene variants in premature newborn infants with severe respiratory distress syndrome. J Perinatol 2018;38:337-44. [Crossref] [PubMed]
- Brasch F, Schimanski S, Mühlfeld C, et al. Alteration of the pulmonary surfactant system in full-term infants with hereditary ABCA3 deficiency. Am J Respir Crit Care Med 2006;174:571-80. [Crossref] [PubMed]
- Guala A, Carrera P, Pastore G, et al. Familial clustering of unexplained transient respiratory distress in 12 newborns from three unrelated families suggests an autosomal-recessive inheritance. Scientific World Journal 2007;7:1611-6. [Crossref] [PubMed]
- Somaschini M, Nogee LM, Sassi I, et al. Unexplained neonatal respiratory distress due to congenital surfactant deficiency. J Pediatr 2007;150:649-53. [Crossref] [PubMed]
- Shen CL, Zhang Q, Meyer J, et al. Genetic Factors Contribute to Risk for Neonatal Respiratory Distress Syndrome among Moderately Preterm, Late Preterm, and Term Infants. J Pediatr 2016;172:69-74.e2. [Crossref] [PubMed]
- Schindlbeck U, Wittmann T, Hoppner S, et al. ABCA3 missense mutations causing surfactant dysfunction disorders have distinct cellular phenotypes. Hum Mutat 2018;39:841-50. [Crossref] [PubMed]
- Bhandari V. Bronchopulmonary dysplasia. New Jersey: Springer, 2016.
- Besnard V, Matsuzaki Y, Clark J, et al. Conditional deletion of ABCA3 in alveolar type II cells alters surfactant homeostasis in newborn and adult mice. Am J Physiol Lung Cell Mol Physiol 2010;298:L646-59. [Crossref] [PubMed]
- Wambach JA, Casey AM, Fishman MP, et al. Genotype-phenotype correlations for infants and children with ABCA3 deficiency. Am J Respir Crit Care Med 2014;189:1538-43. [Crossref] [PubMed]
- Wambach JA, Wegner DJ, Depass K, et al. Single ABCA3 mutations increase risk for neonatal respiratory distress syndrome. Pediatrics 2012;130:e1575-82. [Crossref] [PubMed]
- Khemani RG, Smith LS, Zimmerman JJ, et al. Pediatric acute respiratory distress syndrome: definition, incidence, and epidemiology: proceedings from the Pediatric Acute Lung Injury Consensus Conference. Pediatr Crit Care Med 2015;16:S23-40. [Crossref] [PubMed]
- De Luca D, van Kaam AH, Tingay DG, et al. The Montreux definition of neonatal ARDS: biological and clinical background behind the description of a new entity. Lancet Respir Med 2017;5:657-66. [Crossref] [PubMed]
- Rooth G, Huch A, Huch R. Transcutaneous oxygen monitors are reliable indicators of arterial oxygen tension (if used correctly). Pediatrics 1987;79:283-6. [PubMed]
- Sandberg KL, Brynjarsson H, Hjalmarson O. Transcutaneous blood gas monitoring during neonatal intensive care. Acta Paediatr 2011;100:676-9. [Crossref] [PubMed]
- Li L, Deheragoda M, Lu Y, et al. Hypothyroidism Associated with ATP8B1 Deficiency. J Pediatr 2015;167:1334-9.e1. [Crossref] [PubMed]
- Meng L, Pammi M, Saronwala A, et al. Use of Exome Sequencing for Infants in Intensive Care Units: Ascertainment of Severe Single-Gene Disorders and Effect on Medical Management. JAMA Pediatr 2017;171:e173438. [Crossref] [PubMed]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 2009;4:1073-81. [Crossref] [PubMed]
- Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods 2010;7:248-9. [Crossref] [PubMed]
- Sexton CE, Wadsworth ME, Miller JB, et al. Splice Site Variant Analyzer: Determining the Pathogenicity of Splice Site Variants. J Biomed Res Prac 2018;2:100011.
- Ramani R, Krumholz K, Huang YF, et al. PhastWeb: a web interface for evolutionary conservation scoring of multiple sequence alignments using phastCons and phyloP. Bioinformatics 2019;35:2320-2. [Crossref] [PubMed]
- Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 2015;17:405-24. [Crossref] [PubMed]
- Hansell DM, Bankier AA, Macmahon H, et al. Fleischner Society: glossary of terms for thoracic imaging. Radiology 2008;246:697-722. [Crossref] [PubMed]
- Kröner C, Wittmann T, Reu S, et al. Lung disease caused by ABCA3 mutations. Thorax 2017;72:213-20. [Crossref] [PubMed]
- Shulenin S, Nogee LM, Annilo T, et al. ABCA3 gene mutations in newborns with fatal surfactant deficiency. N Engl J Med 2004;350:1296-303. [Crossref] [PubMed]
- Beers MF, Mulugeta S. The biology of the ABCA3 lipid transporter in lung health and disease. Cell Tissue Res 2017;367:481-93. [Crossref] [PubMed]
- Hallik M, Annilo T, Ilmoja ML. Different course of lung disease in two siblings with novel ABCA3 mutations. Eur J Pediatr 2014;173:1553-6. [Crossref] [PubMed]
- Wambach JA, Yang P, Wegner DJ, et al. Functional Characterization of ATP-Binding Cassette Transporter A3 Mutations from Infants with Respiratory Distress Syndrome. Am J Respir Cell Mol Biol 2016;55:716-21. [Crossref] [PubMed]
- Wittmann T, Frixel S, Hoppner S, et al. Increased Risk of Interstitial Lung Disease in Children with a Single R288K Variant of ABCA3. Mol Med 2016;22:183-91. [Crossref] [PubMed]
- Jackson T, Wegner DJ, White FV, et al. Respiratory failure in a term infant with cis and trans mutations in ABCA3. J Perinatol 2015;35:231-2. [Crossref] [PubMed]
- Wambach J, Tam-Williams JB, Wegner DJ, et al. Multiplexed Direct Genomic Selection to Identify Non-Coding and Copy Number Variation in ABCA3 Among Neonates with Respiratory Failure and Children with Interstitial Lung Disease (chILD) with Monoallelic ABCA3 Variants. Am J Respir Crit Care Med 2018;197:A6323.
- Peca D, Cutrera R, Masotti A, et al. ABCA3, a key player in neonatal respiratory transition and genetic disorders of the surfactant system. Biochem Soc Trans 2015;43:913-9. [Crossref] [PubMed]
(English Language Editors: A. Kassem and J. Reynolds)



