
Influence of KIT mutations on prognosis of pediatric patients with core-binding factor acute myeloid leukemia: a systematic review and meta-analysis
Introduction
Core-binding factor (CBF) is a transcription factor involved in the regulation of hematopoiesis. Mutations in the genes encoding CBF subunits have been found in patients with acute myeloid leukemia (AML). AML with CBF gene mutations (CBF-AML) is characterized by the presence of one of the following two chromosomal rearrangements: t(Jeny8;21)(q22;q22) and inv(Jeny16)(p13;q22)/t(Jeny16;16)(p13;q22), which are usually shortened to t(Jeny8;21) and inv(Jeny16), respectively. The chromosomal anomalies disrupt the encoding of the CBF subunits CBF-α and CBF-β, respectively. Patients with CBF-AML usually have a better prognosis than patients with other AML subtypes (1); however, the outcomes of CBF-AML are heterogeneous. Relapse occurs in ~30% of adult (2) and pediatric CBF-AML patients (3) due to as yet unknown causes.
KIT is a type III tyrosine kinase receptor that is expressed on the surface of hematopoietic stem cells, and KIT signaling is involved in hematopoiesis. Interestingly, mutations in the KIT gene are found in only a small proportion of AML patients (4–5%), but these mutations are present in nearly a third of all CBF-AML patients (approximately 30%) (1,4). The influence of KIT mutations on the prognosis of CBF-AML patients is currently unclear, with some studies reporting that these mutations are unfavorable (5,6) and others reporting no obvious prognostic impact (1,7,8). The 2017 European Leukemia Net (9) and the 2019 National Comprehensive Cancer Network (10) guidelines for AML do not put CBF-AML associated with KIT mutations in the intermediate prognosis category, but only state that the prognosis may be less favorable than that of patients with wild-type KIT genes. At present, there is insufficient data on the prognostic implications of KIT mutations in CBF-AML patients, especially pediatric patients. We therefore carried out a meta-analysis of studies assessing the effects of KIT mutations on the prognosis of pediatric CBF-AML patients. We present the following article in accordance with the PRISMA reporting checklist (available at http://dx.doi.org/10.21037/tp-20-102).
Methods
Literature search
An electronic search of the PubMed, Cochrane Library, and Embase databases was conducted using the following search terms: proto-oncogene proteins c-KIT or KIT or CD117 or Stem cell factor receptor or SCFR or SCF receptor or SCFR; and acute myeloid leukemia or AML or leukemia, myeloid, acute or acute nonlymphocytic leukemia or ANLL. The reference lists of eligible studies were also reviewed to complete the search.
The following inclusion criteria were used for this meta-analysis: (I) original articles, (II) English-language papers, (III) papers published between January 2000 and June 2019, (IV) studies involving pediatric CBF-AML patients with either t(Jeny8.21) or inv(Jeny16) or both types of patients, and (V) studies providing detailed survival information according to the KIT mutational status. We excluded studies that focused on adult CBF-AML patients. In the case of duplicate studies, only one article was considered.
Data extraction and quality assessment
To avoid bias, the eligibility of the studies was independently determined by two co-authors (Junjie Fan and Li Gao), and any disagreements were resolved by consultation with another author (Shaoyan Hu). The two researchers (Junjie Fan and Li Gao) read the full texts of selected articles and extracted the following data from each study: first author’s name, publication year, study location(s), patient age, sample size, sites of mutation on the KIT gene, and clinical outcomes, namely, the rates of complete remission (CR), relapse, overall survival (OS), event-free survival (EFS), and disease-free survival (DFS). If any of the above data points could not be acquired from the published studies, the authors of the article were requested to provide the data. The quality of the included studies was assessed using the Newcastle-Ottawa scale (NOS) (11). NOS scores were independently calculated by two reviewers (Junjie Fan and Jing Chen). NOS scores were categorized as follows: ≥7 points, high quality; 4–6 points, medium quality; and 3 or fewer points, low quality.
Statistical analysis
For this meta-analysis, we used the RevMan version 5.2 and Stata version 12.0 software packages. The CR, relapse, OS, EFS, and DFS rates were calculated for each study. Risk ratios (RRs) were used to assess the probabilities of CR and relapse according to the presence or absence of KIT mutations. The OS, EFS, and DFS rates were evaluated using hazard ratios (HRs). If Kaplan-Meier survival curves had been provided, the HRs were calculated using the Parmar method (12), and the GetData Graph Digitizer software was used to extract data from the survival curves. RRs of >1 signified that the presence of KIT mutations was related with higher rates of CR or relapse, while HRs of >1 signified that the presence of KIT mutations was linked to lower rates of OS, DFS, or EFS.
Initially, the fixed-effects model was used to analyze all data, and then, the I2 statistic was used to identify statistically significant heterogeneity between the studies. Heterogeneity was quantified as mild, moderate, and severe, with upper limits of 25%, 50%, and 75% for I2, respectively. If the I2 value was 50% or lower, we selected the fixed-effects model; if not, we chose the random-effects model (13,14).
We carried out “leave-one-out” sensitivity analysis: one study included in the meta-analysis was evaluated each time to confirm that our findings were not significantly changed by any single study. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Results
Study selection
After the electronic literature search, we removed irrelevant studies by screening the titles and reviewing the abstracts; then, we retrieved the full texts of the remaining 48 articles. We further removed 4 articles that were not written in English and 3 that were conference abstracts without detailed data. After reviewing the references cited within the remaining 41 articles, we added 1 more eligible article.
From the 42 candidate articles, we excluded 22 articles that focused on adult subjects, 4 articles that included both adult and pediatric subjects and did not provide isolated pediatric survival data, 2 duplicate articles with the same study subjects, 1 study with insufficient survival data, and 1 study that focused on experimental methods. Thus, a total of 12 studies were finally included in this meta-analysis (3,7,15-24). A flow chart of the study-selection process is shown in Figure 1.
Study characteristics
The 12 selected studies were published between 2005 and 2019. Six studies had been conducted in Asia, three in Europe, two in America, and one in Egypt. The 12 studies included a total of 1,123 children with CBF-AML, including 826 children with t(Jeny8;21), and 297 children with inv(Jeny16). A total of 256 patients had KIT mutations, and the incidence of KIT mutations varied from 14.7% to 45.3%. The study characteristics have been listed in Table 1.
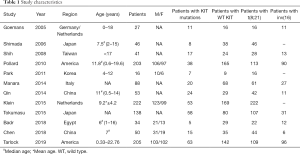
Full table
Of the 12 studies in this meta-analysis, 10 studies had NOS scores of ≥7 and were deemed to be of high quality, and 2 studies had NOS scores of 4–6 and were considered to be of medium quality. The average NOS score was 7.17.
Outcomes of the meta-analysis
We evaluated 5 prognostic indexes to determine the effects of KIT mutations on the clinical outcomes of pediatric CBF-AML patients. Our results are listed below:
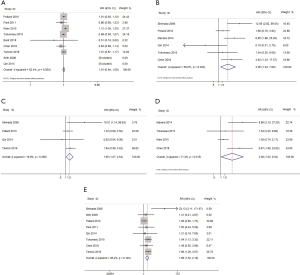
CR rate: 9 studies (3,7,17,18,20-24), 897 patients, pooled RR: 1.01, 95% CI: 0.94–1.09, P=0.761.
OS rate: 6 studies (3,7,16,19,20,22), 528 patients, pooled HR: 3.05, 95% CI: 1.23–7.60, P=0.016.
EFS rate: 4 studies (3,19,21,22), 459 patients, pooled HR: 3.08, 95% CI: 1.02–9.32, P=0.046.
DFS rate: 4 studies (7,16,20,24), 460 patients, pooled HR: 1.65, 95% CI: 1.07–2.54, P=0.024.
Relapse rate: 8 studies (3,7,16-18,20,22,24), 628 patients, pooled RR: 1.69, 95% CI: 1.32–2.16, P=0.000.
The above findings show that the presence of KIT mutations did not affect the CR rate in pediatric CBF-AML patients (P>0.05). However, the presence of KIT mutations was associated with significantly lower OS, DFS, and EFS rates and significantly higher relapse risk in children with CBF-AML (P<0.05 for all; Figure 2). We also performed a sensitivity analysis, and the results indicated that no single study had a significant influence on the pooled RRs and HRs.
Discussion
CBF abnormalities alter cell proliferation and impair the differentiation of hematopoietic progenitor cells, and thereby lead to the development of a leukemogenic profile (25,26). However, these abnormalities by themselves are insufficient to cause leukemia, and the presence of other mutations is necessary for the development of the full leukemic phenotype. KIT is a receptor tyrosine kinase type III expressed on the surface of hematopoietic stem cells. Activation of the KIT receptor results in the phosphorylation of various substrates, thereby activating a signaling cascade and changing the activity of the cell. Recently, KIT mutations were linked to CBF-AML (15,27). KIT mutations can cause over-activation of the KIT receptors, and have been described as the second ‘hit’ in the two-hit model of leukemic transformation (17,28). The relationship between KIT mutations and the prognosis of CBF-AML patients has drawn extensive attention, for example, a study across four pediatric clinical trials found no prognostic significance of KIT mutations, even when accounting for mutational site (7), while some other studies found that KIT mutations were associated with inferior clinical outcomes (3,16). Thus, no clear consensus has emerged so far.
A meta-analysis published in 2016 (29) attempted to clarify this question. It found that KIT mutations had a negative effect on the relapse risk in CBF-AML and t(Jeny8;21) AML, but not on the OS in CBF-AML and the CR, relapse risk, and OS in inv(Jeny16) AML. That meta-analysis focused mainly on adult patients, but the authors found that omitting two studies that focused on pediatric patients did not alter the results or reduce the heterogeneity. Many new studies have been published on this topic in recent years, and to our knowledge, the present meta-analysis is the first to focus on the prognostic implications of KIT mutations in children with CBF-AML. Our study showed that KIT mutations did not affect the CR rate, but did affect more long-term indexes of therapeutic effect, including OS, EFS, DFS, and relapse rate. In many AML protocols, CR as an early treatment response is considered to affect prognosis, and the treatment should be adjusted accordingly. Our study suggests that pediatric CBF-AML patients with KIT mutations should stratified into a higher-risk group although their CR rate was not affected. In addition, other measures, apart from chemotherapy, may be required to avoid these adverse effects. Some preliminary studies have shown that additional interventions may improve the prognosis of this group of patients. Tarlock et al. (24) demonstrated that gemtuzumab ozogamicin and tyrosine kinase inhibitors can improve the prognosis of these patients. Tokumasu et al. (22) also reported that tyrosine kinase inhibitors can reduce the relapse risk, and that subsequent hematologic stem cell transplantation is needed for better treatment outcomes. Thus, for pediatric CBF-AML patients, the detection of KIT mutations should be routine conducted during the early diagnostic workup and risk stratification, and novel therapies apart from the regular chemotherapy should be evaluated in the future.
Our study has certain limitations. First, the sample size was small even after the data from the included studies were pooled. Second, t(Jeny8;21) AML and inv(Jeny16) AML differ in terms of both disease and patient characteristics (4,30-32), and so, the prognostic effects of KIT mutations may also differ between these two AML subtypes. We could not conduct subgroup analyses for these CBF-AML subtypes due to the small sample sizes of the included studies, especially for inv(Jeny16) AML patients. Third, different mutation sites could have different effects on prognosis. For example, the KIT D816 mutation at exon 17, which encodes the activation loop of the tyrosine kinase domain, shows a strong adverse effect on prognosis, while mutations at exon 8 or other sites may have no obvious prognostic impact (33,34). A subgroup analysis based on the functions of the KIT mutations would be useful to resolve this issue, however, most of the included studies did not stratify survival data according to mutation site. In addition, some other factors such as mutant level or methylation level of the KIT gene may also affect its signaling pathway (35,36), and these factors should also be taken into consideration in the future work. Forth, the influence of gene variations may differ in different ethnic groups, and this may partly account for the differences in the results of different studies. However, most of the included studies did not report the race of the patients, and therefore, we could not do a subgroup analysis based on race/ethnicity. Finally, the methods used for disease detection and minimal residual disease detection, and the gene sequencing technology likely varied widely over time and among individual institutions, therefore, the reliability of the data of some of the included studies may be poor, and this may have affected the results of this meta-analysis.
In conclusion, our study suggests that KIT mutations are linked to a poor prognosis in children with CBF-AML. Pediatric patients with these mutations may need to be stratified into a higher risk group, and additional therapies may need to be considered for their treatment.
Acknowledgments
Funding: This research was supported by grants from the National Natural Science Foundation of China (81770193), Jiangsu Innovation Team (CXTDA2017014), and Social Development Project of Jiangsu Province (BE2017659).
Footnote
Reporting Checklist: The authors have completed the PRISMA reporting checklist. Available at http://dx.doi.org/10.21037/tp-20-102
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tp-20-102). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Riera L, Marmont F, Toppino D, et al. Core binding factor acute myeloid leukaemia and c-KIT mutations. Oncol Rep 2013;29:1867-72. [Crossref] [PubMed]
- Jourdan E, Boissel N, Chevret S, et al. Prospective evaluation of gene mutations and minimal residual disease in patients with core binding factor acute myeloid leukemia. Blood 2013;121:2213-23. [Crossref] [PubMed]
- Chen X, Dou H, Wang X, et al. KIT mutations correlate with adverse survival in children with core-binding factor acute myeloid leukemia. Leuk Lymphoma 2018;59:829-36. [Crossref] [PubMed]
- Solh M, Yohe S, Weisdorf D, et al. Core-binding factor acute myeloid leukemia: Heterogeneity, monitoring, and therapy. Am J Hematol 2014;89:1121-31. [Crossref] [PubMed]
- Schnittger S, Kohl TM, Haferlach T, et al. KIT-D816 mutations in AML1-ETO-positive AML are associated with impaired event-free and overall survival. Blood 2006;107:1791-9. [Crossref] [PubMed]
- Mosna F, Papayannidis C, Martinelli G, et al. Complex karyotype, older age, and reduced first-line dose intensity determine poor survival in core binding factor acute myeloid leukemia patients with long-term follow-up. Am J Hematol 2015;90:515-23. [Crossref] [PubMed]
- Pollard JA, Alonzo TA, Gerbing RB, et al. Prevalence and prognostic significance of KIT mutations in pediatric patients with core binding factor AML enrolled on serial pediatric cooperative trials for de novo AML. Blood 2010;115:2372-9. [Crossref] [PubMed]
- Cairoli R, Beghini A, Turrini M, et al. Old and new prognostic factors in acute myeloid leukemia with deranged core-binding factor beta. Am J Hematol 2013;88:594-600. [Crossref] [PubMed]
- Döhner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017;129:424-47. [Crossref] [PubMed]
- Tallman MS, Wang ES, Altman JK, et al. Acute Myeloid Leukemia, Version 3.2019, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2019;17:721-49. [Crossref] [PubMed]
- Wells G, Shea B, O'Connell D, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. Symposium on Systematic Reviews: Beyond the Basics 2014.
- Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med 1998;17:2815-34. [Crossref] [PubMed]
- Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ 2003;327:557-60. [Crossref] [PubMed]
- Melsen WG, Bootsma MC, Rovers MM, et al. The effects of clinical and statistical heterogeneity on the predictive values of results from meta-analyses. Clin Microbiol Infect 2014;20:123-9. [Crossref] [PubMed]
- Goemans BF, Zwaan Ch M, Miller M, et al. Mutations in KIT and RAS are frequent events in pediatric core-binding factor acute myeloid leukemia. Leukemia 2005;19:1536-42. [Crossref] [PubMed]
- Shimada A, Taki T, Tabuchi K, et al. KIT mutations, and not FLT3 internal tandem duplication, are strongly associated with a poor prognosis in pediatric acute myeloid leukemia with t(8;21): A study of the Japanese ChildhoodAMLCooperative Study Group. Blood 2006;107:1806-9. [Crossref] [PubMed]
- Shih LY, Liang DC, Huang CF, et al. Cooperating mutations of receptor tyrosine kinases and Ras genes in childhood core-binding factor acute myeloid leukemia and a comparative analysis on paired diagnosis and relapse samples. Leukemia 2008;22:303-7. [Crossref] [PubMed]
- Park SH, Chi HS, Min SK, et al. Prognostic impact of c-KIT mutations in core binding factor acute myeloid leukemia. Leuk Res 2011;35:1376-83. [Crossref] [PubMed]
- Manara E, Bisio V, Masetti R, et al. Core-binding factor acute myeloid leukemia in pediatric patients enrolled in the AIEOP AML 2002/01 trial: screening and prognostic impact of c-KIT mutations. Leukemia 2014;28:1132-4. [Crossref] [PubMed]
- Qin YZ, Zhu HH, Jiang Q, et al. Prevalence and prognostic significance of c-KIT mutations in core binding factor acute myeloid leukemia: A comprehensive large-scale study from a single Chinese center. Leuk Res 2014;38:1435-40. [Crossref] [PubMed]
- Klein K, Kaspers G, Harrison CJ, et al. Clinical impact of additional cytogenetic aberrations, cKIT and RAS mutations, and treatment elements in pediatric t(8;21)-AML: Results from an international retrospective study by the International Berlin-Frankfurt-Münster Study Group. J Clin Oncol 2015;33:4247-58. [Crossref] [PubMed]
- Tokumasu M, Murata C, Shimada A, et al. Adverse prognostic impact of KIT mutations in childhood CBF-AML: the results of the Japanese Pediatric Leukemia/Lymphoma Study Group AML-05 trial. Leukemia 2015;29:2438-41. [Crossref] [PubMed]
- Badr P, Elsayed GM, Eldin DN, et al. Detection of KIT mutations in core binding factor acute myeloid leukemia. Leuk Res Rep 2018;10:20-5. [Crossref] [PubMed]
- Tarlock K, Alonzo TA, Wang YC, et al. Functional Properties of KIT Mutations are Associated with Differential Clinical Outcomes and Response to Targeted Therapeutics in CBF Acute Myeloid Leukemia. Clin Cancer Res 2019;25:5038-48. [Crossref] [PubMed]
- Hart SM, Foroni L. Core binding factor genes and human leukemia. Haematologica 2002;87:1307-23. [PubMed]
- Speck NA, Gilliland DG. Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer 2002;2:502-13. [Crossref] [PubMed]
- Boissel N, Leroy H, Brethon B, et al. Incidence and prognostic impact of c-Kit, FLT3, and Ras gene mutations in core binding factor acute myeloid leukemia (CBF-AML). Leukemia 2006;20:965-70. [Crossref] [PubMed]
- Wang YY, Zhao LJ, Wu CF, et al. C-KIT mutation cooperates with full-length AML1-ETO to induce acute myeloid leukemia in mice. Proc Natl Acad Sci U S A 2011;108:2450-5. [Crossref] [PubMed]
- Chen W, Xie H, Wang H, et al. Prognostic Significance of KIT Mutations in Core-Binding Factor Acute Myeloid Leukemia: A Systematic Review and Meta-Analysis. PLoS One 2016;11:e0146614. [Crossref] [PubMed]
- Marcucci G, Mrozek K, Ruppert AS, et al. Prognostic factors and outcome of core binding factor acute myeloid leukemia patients with t(8;21) differ from those of patients with inv(16): a Cancer and Leukemia Group B study. J Clin Oncol 2005;23:5705-17. [Crossref] [PubMed]
- Appelbaum FR, Kopecky KJ, Tallman MS, et al. The clinical spectrum of adult acute myeloid leukaemia associated with core binding factor translocations. Br J Haematol 2006;135:165-73. [Crossref] [PubMed]
- Duployez N, Marceau-Renaut A, Boissel N, et al. Comprehensive mutational profiling of core binding factor acute myeloid leukemia. Blood 2016;127:2451-9. [Crossref] [PubMed]
- Kim HJ, Ahn HK, Jung CW, et al. KIT D816 mutation associates with adverse outcomes in core binding factor acute myeloid leukemia, especially in the subgroup with RUNX1/RUNX1T1 rearrangement. Ann Hematol 2013;92:163-71. [Crossref] [PubMed]
- Yui S, Kurosawa S, Yamaguchi H, et al. D816 mutation of the KIT gene in core binding factor acute myeloid leukemia is associated with poorer prognosis than other KIT gene mutations. Ann Hematol 2017;96:1641-52. [Crossref] [PubMed]
- Jang W, Yoon JH, Park J, et al. Significance of KIT exon 17 mutation depends on mutant level rather than positivity in core-binding factor acute myeloid leukemia. Blood Cancer J 2016;6:e387. [Crossref] [PubMed]
- Vozdova M, Kubickova S, Fictum P, et al. Mutation and methylation status of KIT and TP(53) in canine cutaneous and subcutaneous mast cell tumours. Vet Comp Oncol 2020;18:438-44. [Crossref] [PubMed]



