Energy metabolism in neuroblastoma and Wilms tumor
Introduction
The Warburg effect was first described by Otto Warburg in the 1920s, for which he was honored with a Nobel Prize in 1931. Warburg observed that tumor cells favor oxidizing glucose to lactate, rather than to CO2, even with abundant oxygen present (1), which seems paradoxical at first glance. For decades it was assumed that the Warburg effect was provoked by an insufficient supply of blood vessels to the tumor, leading to hypoxia and thereby forcing the tumor cells to adjust to the oxygen shortage by up-regulating glycolysis. However, even with ample oxygen supply, most solid tumors still show increased glycolytic activity and lowered respiration. In clinical diagnostics, the Warburg effect is exploited in the application of positron emission tomography (PET) to detect malignant tumors and metastases by virtue of their higher glucose uptake and retention compared to normal cells.
In the presence of ample glucose, glycolysis generates ATP more rapidly than oxidative phosphorylation (OXPHOS) (2). It is now accepted that enhanced glycolysis is necessary to provide building blocks for anabolic growth (3). Apart from the generation of ATP, intermediates of glycolysis serve as precursors for a variety of biosynthetic pathways necessary for cell proliferation. Thus, glucose metabolism impacts the synthesis of nucleotides, amino acids and fatty acids (2,4,5).
The bulk of cellular energy is normally produced in mitochondria, the cellular organelles that produce and regulate energy in the form of ATP through the five complexes of the OXPHOS system. Mitochondrial energy metabolism includes pyruvate oxidation, the citric acid cycle, OXPHOS and β-oxidation of fatty acids. Mitochondria additionally are associated with several vital cellular functions, including apoptosis, calcium signaling, cellular stress response, and many anabolic and catabolic pathways (6).
In the last decade several laboratories have investigated changes of the OXPHOS system in cancer. We and others were able to show that, depending on the tumor type, cancer cells are frequently characterized by either a general low amount of mitochondria and OXPHOS (7), a specific defect of one OXPHOS complex (8-12) or a combined low level of OXPHOS complexes (13).
Also, the molecular mechanisms leading to the metabolic rewiring of cancer cells have received intense investigation. Those studies demonstrated that genetic alterations of cancer cells frequently affect oncogenes and tumor suppressor genes that regulate cellular metabolism (Figure 1).
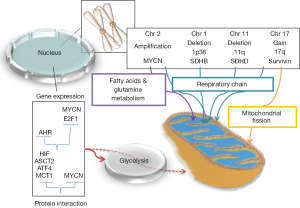
Here we review the literature related to cancer energy metabolism in neuroblastoma (NB) and Wilms tumor (WT). NBs and WTs are the two most frequent tumors in children together accounting for nearly 15% of all pediatric cancers (14,15). They occur in early childhood and typically present as large abdominal masses closely associated with the kidneys. NBs are presumed to originate from neural crest sympathoadrenal progenitors (neuroblasts) and WTs from the primitive metanephrogenic blastema (16).
Regulation of cancer metabolism
The switch to glycolysis and lowered OXPHOS activity in cancer cells can be achieved by a broad variety of mechanisms. Here we provide an overview of the most important regulators of cellular energy metabolism and how oncogenes and tumor suppressors can interact with these regulatory pathways (Figure 1).
The most important sensor of aerobic and anaerobic conditions is hypoxia inducible factor (HIF), which is the major transcription factor responsible for gene expression changes under hypoxic conditions. Tissue hypoxia is involved in the progression and chemoresistance of many types of tumors because it affects a plethora of genes associated with metabolism, angiogenesis, cell survival and apoptosis (17). Under normoxic conditions, HIF-1α undergoes permanent, oxygen-dependent hydroxylation, and is thus recognized and ubiquinated by the von Hippel-Lindau tumor suppressor (VHL) and ultimately degraded. However, HIF-1α can also be activated under normoxic, albeit pathological conditions by certain oncogenic signaling pathways (e.g., mutation of VHL in renal cell carcinoma). In this case a “pseudo-hypoxic” condition is induced while oxygen is present. When activated, HIF-1α stimulates the transcription of genes encoding glucose transporters and many glycolytic enzymes and activates pyruvate dehydrogenase kinases (PDKs), which reduces the flow of glucose-derived pyruvate into the tricarboxylic acid (TCA) cycle (17). AMP-activated protein kinase (AMPK) is a crucial sensor of energy status and plays an important role in the cellular response to metabolic stress. It functions as a metabolic checkpoint, regulating the cellular response to energy availability. When cells undergo energetic stress (owing to prolonged starvation or excessive exercise), AMPK is activated in response to an increased AMP/ATP ratio. Thus, activated AMPK shifts cells to an oxidative metabolic phenotype, provokes cell-cycle arrest and inhibits proliferation (18,19). Cancer cells must escape this checkpoint to proliferate. Several oncogenes are able to suppress AMPK signaling to allow tumor cells to divide even during energetic stress conditions. Loss of AMPK leads to activation of mammalian target of rapamycin (mTOR) and subsequently HIF-1; therefore, inactivation of AMPK contributes to the switch to aerobic glycolysis (20).
The MYC family of genes includes MYC, MYCN and MYCL that encode, respectively, the transcription factors MYC, MYCN and MYCL. Myc transcription factors coordinate the control of cell growth, division, and metabolism (21). MYCs are proto-oncogenes involved in many signal transduction pathways that are responsive to growth factors or to inputs from the cellular microenvironment. Expression of these transcription factors is deregulated in many human cancers. MYC induces an abnormal transcriptional response network, controls cell growth and proliferation, and affects different aspects of cellular metabolism by robustly activating key genes involved in mitochondrial biogenesis, glucose and glutamine metabolism, and lipid synthesis (22). When MYC is highly expressed it collaborates with HIF in the regulation of glucose transporters, glycolytic enzymes, lactate dehydrogenase A (LDHA) and PDK1 (23).
p53 is a tumor suppressor and transcription factor well known for its important roles in apoptosis, the DNA-damage response, and regulation of cellular metabolism. It activates hexokinase 2 (HK2) that converts glucose to glucose-6-phosphate, which enters either glycolysis to produce ATP, or the pentose phosphate pathway to support macromolecule biosynthesis. Under normal conditions, p53 inhibits the glycolytic pathway by supporting the expression of PTEN (phosphatase and tensin homolog) that inhibits phosphatidylinositol 3-kinase (PI3K). Therefore, loss of p53 is an important boost towards acquisition of the glycolytic phenotype (24). Moreover, octamer-binding transcription factor 1 (OCT1) is often overexpressed in several human cancers. OCT1 cooperates with p53 to regulate the balance between glycolytic and oxidative metabolism. OCT1 enhances resistance to oxidative stress, regulates genes responsible for up-regulating glucose metabolism, and reduces mitochondrial respiration in cancer cells (25).
Neuroblastoma (NB)
The beginnings of research on NB go back to a study of 60 cases during 1944-1970 at the M. D. Anderson Hospital and Tumor Institute (26). NB is the most frequent malignancy of the sympathetic nervous system in children, with an incidence of 1.3 cases per 100,000 children (aged 0-14 years) (27,28), accounting for approximately 15% of all cancer deaths in children (28). Histologically and genetically, NB is a heterogeneous tumor with an overall likelihood of 60% that it will metastasize (29). NBs are classified as peripheral neuroblastic tumors (pNTs) that derive from the neural crest sympathoadrenal lineage that would normally develop and differentiate into sympathetic ganglia, chromaffin cells of the adrenal medulla, and the paraganglia (28). Extensive genetic studies on NBs uncovered a variety of genetic changes that can be broadly grouped as chromosomal aberrations and genetic mutations (Figure 1). The most frequent chromosomal aberrations are allelic loss of 1p36 and 11q (30-33) and gain of 17q (34,35). The loss of heterozygosity (LOH) of 1p36 is highly associated with MYCN amplification. Amplification of MYCN is a well-recognized genetic marker of high stage NB with poor prognosis. Somatic mutation or amplification of the anaplastic lymphoma receptor tyrosine kinase (ALK) gene is another frequent genetic lesion in NB (36).
Alterations of the OXPHOS system in neuroblastic tumors
NBs, like most other solid tumors, show high glucose uptake on PET examination (37,38). The Warburgian tendency to glycolysis brings about an elevated need for glucose, and excessive lactate production (6). Accordingly, NBs show a high rate of lactic acid production and a low rate of oxygen consumption, reflecting the switch from mitochondrial OXPHOS to glycolysis (39).
There is strong evidence that the Warburg effect in NBs is not caused by a low mitochondrial mass per cell. Electron microscopy showed numerous round and oval or elongated mitochondria in NB cells (40). Consistent with this observation, we recently reported equal levels of citrate synthase activity and porin expression in human NB samples and normal kidney tissue, indicating a similar mitochondrial mass (41). Furthermore, MitoTracker Green analysis indicated similar amounts of mitochondria in NBs, cerebral cortex and retinal ganglion cells (42). Strikingly, a significant reduction in mtDNA copy number and an overall reduction of OXPHOS enzyme activities were observed in human NB tissue; in comparison with cortical kidney tissue, the activity of succinate dehydrogenase (SDH; or complex II) was most affected (41). Interestingly, a similar phenotype was observed in adult renal cell carcinoma which is characterized by different genetic alterations (7).
Ganglioneuromas (GNs) and ganglioneuroblastomas (GNBs), like NBs, are classified as pNTs that originate from immature sympathetic neuroblasts. Intermixed GNBs are histologically and clinically situated between NBs and the benign GNs, while nodular GNBs are composed of nodules of NB cells in the background of intermixed GNBs. According to the Shimada classification, GNs are Schwannian stroma-dominant tumors, whereas NBs are considered as Schwannian stroma-poor neoplasias (43). Surprisingly, GNs show a severe loss of mitochondria, as indicated by loss of porin and significant reduction of citrate synthase; therefore, their loss of all OXPHOS complexes compared to normal adrenal gland and kidney cortex tissue is likely secondary to mitochondrial loss (44).
The nexus between common genetic alterations in NB and energy metabolism
Deletions of chromosome 1p36 occur in approximately 20-40% of primary NBs. As noted above, LOH of 1p is also highly correlated with MYCN amplification (32,33). Succinate dehydrogenase subunit B (SDHB) maps to chromosome 1p36, a region of frequent LOH in NB (31). There may be a correlation between 1p36 LOH and the low activity of SDH reported by our laboratory (41). Sequencing of all four subunits of SDH and two assembly factors showed no pathological mutations in human NB samples (41). These findings are in agreement with those of two other studies, which also failed to detect pathogenic mutations in SDHB and SDHB in NB. Furthermore, epigenetic studies also could not demonstrate evidence for silencing of SDHB expression (45,46). However, rare cases of NB with SDHB and SDHD mutations have been reported (46-48). NB cells transfected with different SDHB-mutated constructs showed down-regulation of mitochondrial and cytoplasmic metabolism as well as higher growth rates (49).
Deletion of the long arm of chromosome 11 is observed in high-risk NBs with an unfavorable prognosis (33). The SDHD gene is located on 11q23. A significant reduction in SDHD mRNA expression in NBs compared to both normal fetal neuroblasts and various normal adult tissues was demonstrated (46).
Gain of chromosome 17 is also a frequent chromosomal abnormality detected in advanced-stage NBs. Two patterns of gain are seen for this chromosome: either the whole chromosome 17 or only 17q is multiplied (34). NBs with a 17q gain overexpress BIRC5/Survivin, an anti-apoptotic protein (50). It was reported that the survivin protein is involved in reprogramming metabolism in NB cells by inducing fragmentation of mitochondria and reducing mitochondrial respiration. Survivin shifts the energy production from OXPHOS to aerobic glycolysis through inhibition of complex I and induction of mitochondrial fission via recruitment of dynamin 1-like protein/dynamin-related protein 1 (DNM1L/Drp1). NB cells with elevated survivin levels showed faster glucose metabolism and higher lactate production. Furthermore, inhibition of complex I by survivin prevented reactive oxygen species (ROS) accumulation and Forkhead box O3 (FOXO3) activity (50). Feichtinger et al. reported low complex I activity in NBs, although no data regarding gain of chromosome 17 were available (41).
Amplification of MYCN is found in half of all high-risk NBs with poor prognosis and resistance to therapy (51). Studies showed that MYCN is associated with regulation of energy metabolism through direct or indirect activation of genes involved in glycolysis, glutamine metabolism, fatty acids metabolism and mitochondrial function (52-54). Amplification of MYCN supports the Warburg effect in NBs by activating the transcription of several glycolytic genes. Knock-down of MYCN expression in MYCN-amplified NB cell lines caused down-regulation of glycolytic metabolism. Depletion of MYCN led to lower expression of the genes encoding glucose transporter 1 (GLUT1), HK2, PDK1, phosphoglycerate kinase 1 (PGK1), aldolase A (ALDA), and LDHA and leads to decreased proliferation of MYCN-amplified NBs (52).
Interestingly, mitochondrial inhibition by metaiodobenzylguanidine (MIBG) caused destabilization of MYCN and MYC in MYCN-amplified and non-MYCN-amplified NB cells and suppressed NB cell growth (55). MIBG also reduced the ATP to ADP ratio in NB cells (56).
Abundant glutamine is necessary for the TCA cycle in MYCN-amplified NB. Deprivation of glutamine led to depletion of TCA-cycle intermediates (54). During anaplerosis, mitochondrial glutamine metabolism can replenish oxaloacetate (OAA) for the TCA cycle (57). Provision of sufficient levels of glutamine for NB cells relies on activation of amino-acid transporter ASCT2, a glutamine transporter. High expression of MYCN, ATF4 and ASCT2 was observed in MYCN-amplified NB. The elevated level of ASTC2 expression observed in high-stage NB correlates with MYCN and ATF4 expression that promotes aggressive NB progression. Knockdown of MYCN led to a significant reduction in ASCT2 abundance (54).
The expression of the aryl hydrocarbon receptor (AHR), a receptor for dioxin-like compounds, correlates with a higher degree of differentiation of NBs. An inverse correlation between AHR and MYCN levels was reported (58). Members of the E2F family are transcription factors that regulate cell-cycle progression and influence MYCN expression in NBs (59). Ectopic expression of AHR correlates with down-regulation of MYCN and E2F1 expression in NB cells. Silencing of AHR enhanced the expression of both E2F1 and MYCN in NB cells. It is proposed that AHR is an upstream regulator of MYCN expression (58). It was further suggested that AHR and E2F1 are involved in mitochondrial function. Interaction between AHR and ATP5α1, a subunit of complex V, was reported, from which the authors hypothesized that AHR contributes to homeostasis of mitochondrial function and cellular energy metabolism (60). Knockdown of E2F1 in HeLa cells produced highly elevated mRNA levels of genes involved in mitochondrial biogenesis (61). Concurrent findings also support a role for E2F1 in regulating energy homeostasis and mitochondrial function: E2F1 causes a shift from oxidative to glycolytic metabolism under stress conditions (62).
The monocarboxylate transporter (MCT) family member MCT1 can transport both lactate and pyruvate. High activity of MCT1 was correlated with MYCN amplification in NBs. Inhibition of MCT1 causes acidosis and a significant reduction in viability of NB cells (63).
HIF-1α was shown to be expressed in both MYCN-amplified and non-amplified NBs. It seems HIF-1α in association with MYCN fortifies aerobic glycolysis in NBs. Knockdown of HIF1-α selectively decreased the expression of genes encoding GAPDH, HK2, PDK1, ALDA and LDHA in NBs. Concurrent knockdown of both MYCN and HIF-1α further reduced HK2 and LDHA expression (52).
Other regulators of metabolism in NB
Transient receptor potential cation channel, subfamily M, member 2 (TRPM2) is an ion channel that is frequently expressed in cancers. Full-length TRPM2 (TRPM2-L) protected NBs from moderate oxidative stress. The other form of TRPM2 is a dominant-negative short isoform (TRPM2-S). NBs which expressed TRPM2-S showed a significant reduction in tumor growth compared to TRPM2-L-expressing NBs. Significantly reduced expression of HIF-1/2α was detected in TRPM2-S-expressing NB cells. Interestingly, the target proteins regulated by HIF-1/2α in mitochondria (e.g., BNIP3, NDUFA4L2, COX4.1, and COX4.2) were significantly down-regulated in these cells (64).
It was demonstrated that HIF-2α also can play an indirect role in NB metabolism. Over-expressed HIF-2α but weak HIF-1α expression was detected in well-vascularized areas of NBs (65,66). Vascularization, which enables access to more oxygen and metabolites like glucose, glutamine, etc., is necessary for tumor cells. Moreover, blood vessels help tumor cells to get rid of metabolic waste products.
Contingency theories
Accumulation of succinate, the substrate of SDH, correlates with stability of HIF-1α via inhibition of the HIF-alpha prolyl hydroxylases activity (67). Therefore, it is possible that a low level of SDH activity may intensify the hypoxia response and induce reprogramming of metabolism via HIF-1α in NBs. It was shown that hypoxia induces dedifferentiation and a shift to an immature neural crest-like phenotype in NBs, a change associated with greater aggressiveness and a higher potential to metastasize (68).
Wilms tumor (WT)
WT, also known as nephroblastoma, is a pediatric cancer that affects the kidneys (69). The name derives from Dr. Max Wilms, the German surgeon who first described it. Most WTs are unilateral; both kidneys are affected in only 5-6% of cases (70,71). WT usually presents as a single mass encapsulated in fibrous tissue, and thus is well separated from the adjacent kidney tissue (71).
WTs are believed to arise from the malignant transformation of renal stem cells that abnormally persist after embryogenesis and maintain embryonic differentiation capacity. WTs are usually triphasic, which implies they are composed of three main types of tissue: mesenchyma, blastema and epithelium. The mesenchymal tissue is comprised of immature embryonal mesenchyme, but may also contain more differentiated structures (e.g., skeletal muscle, adipose tissue). The blastemal tissue consists of sheets of undifferentiated cells. Finally, WT contains epithelial structures including immature glomeruloid bodies or tubules (71). The overall survival rate of patients with WTs is greater than 90% (15). WTs are histologically divided into two main groups: anaplastic WTs (approximately 10%), characterized by large cells with large hyperchromatic nuclei, multipolar mitotic figures associated with poor response to chemotherapy and lower survival rate; and non-anaplastic WTs that have more favourable survival outcomes (70).
WTs are characterized by genetic abnormalities involving tumor suppressor genes (e.g., WT1 and TP53), oncogenes (e.g., HIF-1 and MYC), and other genes related to the WNT signaling pathway. Recently, somatic mutations in DROSHA and DICER1 were identified in 12% of WTs, two factors regulating miRNA expression (72,73). Furthermore, several genetic syndromes are associated with an increased risk of developing WT. The most common of these are: Wilms-aniridia-genitourinary-mental retardation (WAGR) syndrome, due to deletion of chromosome 11p13 in a region containing the WT1 and PAX6 genes (74,75); Denys-Drash syndrome (DDS), associated with constitutional abnormalities of the WT1 gene (76,77); Beckwith-Wiedemann syndrome (BWS), a pediatric overgrowth disorder with a predisposition to develop embryonal malignancies (78), caused by mutation or deletion of imprinted genes on chromosome 11 (79); and Fanconi anemia (FA), a rare recessive disorder caused by genetic defects in a number of proteins involved in, and associated with chromosomal fragility and genomic instability. There are different subgroups of FA; in FA complementation group D1, a severe form of the disease, biallelic mutation of BRCA2 is causative, and 63% of children with this genotype will develop WTs by the age of 6.7 years (80,81).
Alterations of the energy metabolism in WT
The role of fluoro-D-glucose (FDG)-PET in WTs has not been well established (82). However, there are some case reports demonstrating the WTs can be monitored by PET examination (83). In classical triphasic WT, the epithelial, blastemal and stromal elements differ in terms of mitochondrial energy metabolism. Normal mitochondrial mass was reported for the blastemal and epithelial areas. In contrast, the stroma regions showed severe loss of mitochondria compared to normal kidney tissue, resulting in loss of all OXPHOS complexes, as indicated by low porin expression and citrate synthase activity. The mtDNA copy number was also reduced in WT, which can partly explain the reduction in OXPHOS enzymes (84). These observations are supported by ultrastructural studies, which showed that mesenchymal cells contain few mitochondria, whereas primitive epithelial cells possess numerous mitochondria (85).
In agreement with these data, Hammer and colleagues detected a general decrease of mitochondrial metabolism in WTs. By mass spectrometric analyses of formalin-fixed paraffin-embedded tissue sections, they showed that proteins involved in OXPHOS and other mitochondrial functions (OXPHOS complex components and TCA cycle enzymes) were present at lower levels in WT samples compared with normal kidney tissue. In general, many other metabolic proteins were found to be decreased in WTs compared to normal tissue, for example aldehyde dehydrogenases and proteins related to propanoate and butanoate metabolism, long-chain fatty acid metabolism, and degradation of the branched-chain amino acids valine, leucine and isoleucine. However, because the tissue samples were obtained after chemotherapy treatment, it is unclear if these changes are also present in primary, untreated WT (86).
In WTs, HIF-1α has been found to be expressed at higher levels compared to normal kidney. Moreover, localization of HIF-1α in the tumor tissue is predominantly nuclear, whereas in normal kidney it is cytoplasmic (87,88). As previously mentioned, HIF-1α plays an important role in tumor development in different ways. For example, HIF-1α increases the expression and activity of many different glycolytic protein isoforms, including the transporters GLUT1 and GLUT3 that transport glucose across the plasma membrane, enhancing tumor glycolytic flux. HIF-1α seems also to be involved in the modulation of mitochondrial function and oxygen consumption by inactivating the pyruvate dehydrogenase (PDH) complex in some tumor types (89). Therefore, overexpression of HIF-1α and its nuclear localization, together with the reductions in mitochondrial biogenesis and/or OXPHOS activity found in WTs, suggest that these types of tumors are characterized by the metabolic switch to aerobic glycolysis.
Interestingly, there are no further studies available which have investigated the metabolism of WT in detail regarding the metabolic pathways known to be altered in a range of cancers (see “Regulation of cancer metabolism”). There is only indirect evidence from the genes altered in WT that there are alterations of cell metabolism.
The nexus between genetic alterations in WT and energy metabolism
About 5-6% of WTs present with TP53 mutations. Interestingly, TP53 mutations are mostly found in anaplastic WT, indicating an association with a more aggressive tumor phenotype (58,90,91). TP53 is a tumor suppressor gene involved in the development of many types of cancer. It plays a role in the regulation of many genes, including acting as a transcriptional repressor of GLUT1, and therefore it is important in glycolysis regulation. In anaplastic WT a stronger expression of GLUT1 compared to non-anaplastic WT was observed (92). Moreover, as previously mentioned, GLUT1 is stimulated by HIF-1α. Since p53 mediates the repression of HIF-1α, HIF-1α and p53 seem to cooperate in establishing the Warburg effect in anaplastic WTs and thereby could be responsible for the development of an aggressive phenotype. Interestingly, Zawacka-Pankau et al. showed that pharmacological re-activation of p53 in tumor cell lines and xenografts decreases the levels of HIF-1α, both in normoxic and hypoxic conditions, and they also found that p53 binds to the promoter of HIF-1α, inhibiting its expression. Moreover, they showed that the pharmacological re-activation of p53 is able to down-regulate metabolic genes, including the gene responsible for the transcription of GLUT1, confirming the interdependence of p53 and HIF-1α in the regulation of the Warburg effect in cancer cells (93). The morphology and physiology of WTs resemble those of the developing kidney (94). For example, the WNT pathway is required during embryonic development to maintain the pluripotency and plasticity of human embryonic stem cells (71). Activation of the WNT/β-catenin pathway is an important hallmark of stemness of cancer stem cells, and β-catenin mutations have been found in 15% of WTs (95,96). In WTs, mutations affecting CTNNB1, the gene encoding β-catenin, usually result in constitutive activation of the WNT/β-catenin signaling pathway (95). In one study, the authors reported that at least 50% of tumors with WT1 mutations also had a β-catenin mutation (97,98).
Recently, it was shown that WNT signaling works in a context and tissue specific manner. So far 19 WNT genes were identified. WNT signaling can be divided into a canonical and a non-canonical pathway, which is β-catenin independent. In a recent paper it was shown that WNT3A (non-canonical) can activate mitochondrial biogenesis and oxygen consumption in mouse myotubes. WNT5A suppressed this action (99). In contrast, WNT3A was shown to increase glycolytic capacity during osteoclast differentiation (100). In addition, WNT5A expression correlated with good or bad prognosis depending on the analyzed tumor type (101-104). Activation of the canonical WNT/β-catenin signaling was shown to increase aerobic glycolysis through suppression of mitochondrial respiration (105,106).
An important downstream target of the WNT/β-catenin pathway is MYC, a proto-oncogene that functions as transcription factor for many genes involved in cell cycle. The MYC and MYCN genes have similar 5' noncoding exons and extensive homology in the coding regions, and they are both overexpressed in many WTs (107,108). As MYC promotes glycolysis and energy production and is a key regulator of glutaminolysis in transformed cells (109), and MYCN in association with HIF-1α increases the transcription of glycolytic enzymes (52), these observations indicate that the WNT signaling pathway influences cancer cell metabolism.
Expression of IGF-2 is increased in 69% of WTs (110). IGF-2 is an imprinted gene normally expressed only from the paternal allele. In sporadic WTs, this gene often exhibits bi-allelic expression (111). An increase of IGF-1 or IGF-2 leads to activation of the insulin signaling pathway. A cascade of events leads to the activation of the PI3K-mitogen activated protein kinase (MAPK)-Akt signaling pathway that results in an increase in glucose uptake and metabolism. Furthermore, activation of Akt signaling via down-regulation of PGC-1α decreases mitochondrial biogenesis (6). WT1, located on chromosome 11p13, plays an important role in genitourinary development, encoding a zinc finger transcription factor and acting as a tumor suppressor. WT1 loss-of-function mutations occur in 10-20% of WT cases (95,97). WTX, located on chromosome Xq11.1, was also found to be involved in WT development. The prevalence of WTX mutations is approximately 18% in WTs (112). Both WT1 and WTX mutations cause dysregulation of the WNT/β-catenin pathway, and consequently abnormal cell proliferation (97,112).
Mutations in the WT1 gene can affect E-cadherin expression. It can induce epithelial differentiation during mesenchymal-epithelial transition and E-cadherin expression in the embryonic kidney (113). Loss or reduction of mtDNA results in hypermethylation of the E-cadherin gene promoter (114). When the tumor cell lines, LNrho0-8 (derived from the LNCaP prostate cancer cell line) and MCFrho0 (derived from the MCF-7 breast cancer cell line) were depleted of mtDNA by treatment with ethidium bromide, the cells lost their epithelial features and gained a mesenchymal phenotype. Furthermore, repression of E-cadherin and induction of vimentin expression were observed. Down-regulation of E-cadherin in LNrho0-8 and MCFrho0 cells was shown to be due to hypermethylation of the E-cadherin promoter region (115). We propose that the reduction of mtDNA copy number leading to a decline in OXPHOS activity in WT induces epigenetic changes in nuclear DNA, influencing EMT. Because only 10-15% of sporadic (116,117) WTs carry a WT1 mutation, but many have low mtDNA levels, low mtDNA and OXPHOS might be the main trigger for low E-cadherin expression.
It has been shown in human embryonic kidney cells and murine mesonephric cells, that inducing or repressing WT1 expression affects the expression of genes involved in the synthesis of cholesterol and fatty acids (118). The mevalonate pathway is involved in the production of cholesterol, vitamin D, steroid hormones and bile acids. Transcription of these genes needs activation by sterol regulatory element-binding proteins (SREBPs). Rae et al. hypothesize that WT1 interacts with SREBP to block transcription of mevalonate pathway genes (118). Therefore, it is reasonable to speculate that, in tumors where WT1 is mutated, the synthesis of cholesterol and other fatty acids is increased. This would offer an advantage to the tumor since these are building blocks for cellular membranes and thus important for cell proliferation.
Therapeutic options
As previously mentioned, NB and WT cells demonstrate high glucose uptake and preferentially ferment glucose to lactate instead to CO2 and H2O (OXPHOS), which shows their dependency on glycolysis (83). In addition, it was discussed that NBs and WTs have OXPHOS deficiencies. All this evidence supports the idea that targeting the Warburg effect may be a promising therapeutic strategy against NB and WT. Some results that support this idea in NBs are presented next.
Inhibition of glycolysis causes a decrease in NB cell proliferation in vitro, underlining the reliance of NB cells on glycolysis. A xenograft study showed that 3-BrOP, a novel inhibitor of glycolysis, was able to decrease NB mass by 78% in mice (39). Dichloroacetate (DCA) is a drug which indirectly activates PDH, causing a shift in pyruvate metabolism from lactate to the TCA cycle. It was shown that DCA is able to reduce lactate production and enhance oxygen consumption in NB cells and inhibits proliferation of highly malignant NB cells (119,120). Inhibition of LDHA, the enzyme that metabolizes pyruvate to lactate, was also able to suppress the tumorigenic capacity of NBs in vivo (52). Depriving NBs of glucose had detrimental effects on them. Glucose-free medium supplemented with the ketone bodies, acetoacetate and hydroxybutyrate reduced the viability of NB cells (121). Also, a higher level of NB cell differentiation was observed in low-glucose medium (122). Stimulating NB cells to differentiate by treating them with differentiating agents such retinoic acid (RA) or 2-O, 3-O-desulfated heparin (ODSH) had a suppressive effect on tumor growth and metastasis (123,124). As NB cells are sensitive to glucose depletion, a high-fat low-carbohydrate ketogenic diet can be considered a non-toxic therapeutic strategy. On a ketogenic diet, NB cells would be forced to obtain their energy from fatty acids through OXPHOS. However, since NBs are unable to respire due to mitochondrial defects, they might be selectively killed by a ketogenic diet.
Acknowledgements
Funding: This work was supported by the Vereinigung zur Förderung der pädiatrischen Forschung und Fortbildung Salzburg, Children Cancer Foundation, the Austrian Research Promotion Agency (822782/THERAPEP) and the Mitochondrial European Educational Training (MEET), ITN MARIE CURIE PEOPLE (317433).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Warburg OH. eds. The Metabolism of Tumours: Investigations from the Kaiser Wilhelm Institute for Biology, Berlin-Dahlem. New York: Richard R. Smith, 1931:129-69.
- Wu W, Zhao S. Metabolic changes in cancer: beyond the Warburg effect. Acta Biochim Biophys Sin (Shanghai) 2013;45:18-26. [PubMed]
- Koppenol WH, Bounds PL, Dang CV. Otto Warburg's contributions to current concepts of cancer metabolism. Nat Rev Cancer 2011;11:325-37. [PubMed]
- Soga T. Cancer metabolism: key players in metabolic reprogramming. Cancer Sci 2013;104:275-81. [PubMed]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 2009;324:1029-33. [PubMed]
- Wallace DC. Mitochondria and cancer. Nat Rev Cancer 2012;12:685-98. [PubMed]
- Meierhofer D, Mayr JA, Foetschl U, et al. Decrease of mitochondrial DNA content and energy metabolism in renal cell carcinoma. Carcinogenesis 2004;25:1005-10. [PubMed]
- Zimmermann FA, Mayr JA, Feichtinger R, et al. Respiratory chain complex I is a mitochondrial tumor suppressor of oncocytic tumors. Front Biosci (Elite Ed) 2011;3:315-25. [PubMed]
- Calabrese C, Iommarini L, Kurelac I, et al. Respiratory complex I is essential to induce a Warburg profile in mitochondria-defective tumor cells. Cancer Metab 2013;1:11. [PubMed]
- Astuti D, Douglas F, Lennard TW, et al. Germline SDHD mutation in familial phaeochromocytoma. Lancet 2001;357:1181-2. [PubMed]
- Bayley JP, Oldenburg RA, Nuk J, et al. Paraganglioma and pheochromocytoma upon maternal transmission of SDHD mutations. BMC Med Genet 2014;15:111. [PubMed]
- Lefebvre M, Foulkes WD. Pheochromocytoma and paraganglioma syndromes: genetics and management update. Curr Oncol 2014;21:e8-e17. [PubMed]
- Feichtinger RG, Weis S, Mayr JA, et al. Alterations of oxidative phosphorylation complexes in astrocytomas. Glia 2014;62:514-25. [PubMed]
- Davidoff AM. Neuroblastoma. Semin Pediatr Surg 2012;21:2-14. [PubMed]
- Davidoff AM. Wilms' tumor. Curr Opin Pediatr 2009;21:357-64. [PubMed]
- Marshall GM, Carter DR, Cheung BB, et al. The prenatal origins of cancer. Nat Rev Cancer 2014;14:277-89. [PubMed]
- Kaelin WG Jr. The von Hippel-Lindau tumour suppressor protein: O2 sensing and cancer. Nat Rev Cancer 2008;8:865-73. [PubMed]
- Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer 2011;11:85-95. [PubMed]
- Jones RG, Plas DR, Kubek S, et al. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell 2005;18:283-93. [PubMed]
- Wang W, Guan KL. AMP-activated protein kinase and cancer. Acta Physiol (Oxf) 2009;196:55-63. [PubMed]
- Wahlström T, Arsenian Henriksson M. Impact of MYC in regulation of tumor cell metabolism. Biochim Biophys Acta 2014. [Epub ahead of print]. [PubMed]
- Dang CV. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb Perspect Med 2013.3. [PubMed]
- Dang CV, Kim JW, Gao P, et al. The interplay between MYC and HIF in cancer. Nat Rev Cancer 2008;8:51-6. [PubMed]
- Matoba S, Kang JG, Patino WD, et al. p53 regulates mitochondrial respiration. Science 2006;312:1650-3. [PubMed]
- Almeida R, Almeida J, Shoshkes M, et al. OCT-1 is over-expressed in intestinal metaplasia and intestinal gastric carcinomas and binds to, but does not transactivate, CDX2 in gastric cells. J Pathol 2005;207:396-401. [PubMed]
- Knudson AG Jr, Strong LC. Mutation and cancer: neuroblastoma and pheochromocytoma. Am J Hum Genet 1972;24:514-32. [PubMed]
- Schwab M, Westermann F, Hero B, et al. Neuroblastoma: biology and molecular and chromosomal pathology. Lancet Oncol 2003;4:472-80. [PubMed]
- Park JR, Eggert A, Caron H. Neuroblastoma: biology, prognosis, and treatment. Hematol Oncol Clin North Am 2010;24:65-86. [PubMed]
- Cheung NK, Dyer MA. Neuroblastoma: developmental biology, cancer genomics and immunotherapy. Nat Rev Cancer 2013;13:397-411. [PubMed]
- Maris JM, Weiss MJ, Guo C, et al. Loss of heterozygosity at 1p36 independently predicts for disease progression but not decreased overall survival probability in neuroblastoma patients: a Children's Cancer Group study. J Clin Oncol 2000;18:1888-99. [PubMed]
- Martinsson T, Sjöberg RM, Hallstensson K, et al. Delimitation of a critical tumour suppressor region at distal 1p in neuroblastoma tumours. Eur J Cancer 1997;33:1997-2001. [PubMed]
- Carén H, Fransson S, Ejeskär K, et al. Genetic and epigenetic changes in the common 1p36 deletion in neuroblastoma tumours. Br J Cancer 2007;97:1416-24. [PubMed]
- Guo C, White PS, Weiss MJ, et al. Allelic deletion at 11q23 is common in MYCN single copy neuroblastomas. Oncogene 1999;18:4948-57. [PubMed]
- Plantaz D, Mohapatra G, Matthay KK, et al. Gain of chromosome 17 is the most frequent abnormality detected in neuroblastoma by comparative genomic hybridization. Am J Pathol 1997;150:81-9. [PubMed]
- Theissen J, Oberthuer A, Hombach A, et al. Chromosome 17/17q gain and unaltered profiles in high resolution array-CGH are prognostically informative in neuroblastoma. Genes Chromosomes Cancer 2014;53:639-49. [PubMed]
- Maris JM. Recent advances in neuroblastoma. N Engl J Med 2010;362:2202-11. [PubMed]
- Shulkin BL, Mitchell DS, Ungar DR, et al. Neoplasms in a pediatric population: 2-[F-18]-fluoro-2-deoxy-D-glucose PET studies. Radiology 1995;194:495-500. [PubMed]
- Freebody J, Wegner EA, Rossleigh MA. 2-deoxy-2-((18)F)fluoro-D-glucose positron emission tomography/computed tomography imaging in paediatric oncology. World J Radiol 2014;6:741-55. [PubMed]
- Levy AG, Zage PE, Akers LJ, et al. The combination of the novel glycolysis inhibitor 3-BrOP and rapamycin is effective against neuroblastoma. Invest New Drugs 2012;30:191-9. [PubMed]
- Lyser KM. Low- and high-voltage electron microscopy of a human neuroblastoma in long-term organ culture. Cancer Res 1974;34:594-602. [PubMed]
- Feichtinger RG, Zimmermann F, Mayr JA, et al. Low aerobic mitochondrial energy metabolism in poorly- or undifferentiated neuroblastoma. BMC Cancer 2010;10:149. [PubMed]
- Hoegger MJ, Lieven CJ, Levin LA. Differential production of superoxide by neuronal mitochondria. BMC Neurosci 2008;9:4. [PubMed]
- Shimada H, Ambros IM, Dehner LP, et al. The International Neuroblastoma Pathology Classification (the Shimada system). Cancer 1999;86:364-72. [PubMed]
- Feichtinger RG, Neureiter D, Mayr JA, et al. Loss of mitochondria in ganglioneuromas. Front Biosci (Elite Ed) 2011;3:179-86. [PubMed]
- Astuti D, Morris M, Krona C, et al. Investigation of the role of SDHB inactivation in sporadic phaeochromocytoma and neuroblastoma. Br J Cancer 2004;91:1835-41. [PubMed]
- De Preter K, Vandesompele J, Hoebeeck J, et al. No evidence for involvement of SDHD in neuroblastoma pathogenesis. BMC Cancer 2004;4:55. [PubMed]
- Armstrong R, Greenhalgh KL, Rattenberry E, et al. Succinate dehydrogenase subunit B (SDHB) gene deletion associated with a composite paraganglioma/neuroblastoma. J Med Genet 2009;46:215-6. [PubMed]
- Schimke RN, Collins DL, Stolle CA. Paraganglioma, neuroblastoma, and a SDHB mutation: Resolution of a 30-year-old mystery. Am J Med Genet A 2010;152A:1531-5. [PubMed]
- Rapizzi E, Ercolino T, Fucci R, et al. Succinate dehydrogenase subunit B mutations modify human neuroblastoma cell metabolism and proliferation. Horm Cancer 2014;5:174-84. [PubMed]
- Hagenbuchner J, Kuznetsov AV, Obexer P, et al. BIRC5/Survivin enhances aerobic glycolysis and drug resistance by altered regulation of the mitochondrial fusion/fission machinery. Oncogene 2013;32:4748-57. [PubMed]
- Brodeur GM, Seeger RC, Schwab M, et al. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science 1984;224:1121-4. [PubMed]
- Qing G, Skuli N, Mayes PA, et al. Combinatorial regulation of neuroblastoma tumor progression by N-Myc and hypoxia inducible factor HIF-1alpha. Cancer Res 2010;70:10351-61. [PubMed]
- Zirath H, Frenzel A, Oliynyk G, et al. MYC inhibition induces metabolic changes leading to accumulation of lipid droplets in tumor cells. Proc Natl Acad Sci U S A 2013;110:10258-63. [PubMed]
- Ren P, Yue M, Xiao D, et al. ATF4 and N-Myc coordinate glutamine metabolism in MYCN-amplified neuroblastoma cells through ASCT2 activation. J Pathol 2015;235:90-100. [PubMed]
- Wang SS, Hsiao R, Limpar MM, et al. Destabilization of MYC/MYCN by the mitochondrial inhibitors, metaiodobenzylguanidine, metformin and phenformin. Int J Mol Med 2014;33:35-42. [PubMed]
- Cornelissen J, Wanders RJ, Van den Bogert C, et al. Meta-iodobenzylguanidine (MIBG) inhibits malate and succinate driven mitochondrial ATP synthesis in the human neuroblastoma cell line SK-N-BE(2c). Eur J Cancer 1995;31A:582-6. [PubMed]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, et al. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 2008;7:11-20. [PubMed]
- Wu PY, Liao YF, Juan HF, et al. Aryl hydrocarbon receptor downregulates MYCN expression and promotes cell differentiation of neuroblastoma. PLoS One 2014;9:e88795. [PubMed]
- Strieder V, Lutz W. E2F proteins regulate MYCN expression in neuroblastomas. J Biol Chem 2003;278:2983-9. [PubMed]
- Tappenden DM, Lynn SG, Crawford RB, et al. The aryl hydrocarbon receptor interacts with ATP5α1, a subunit of the ATP synthase complex, and modulates mitochondrial function. Toxicol Appl Pharmacol 2011;254:299-310. [PubMed]
- Goto Y, Hayashi R, Kang D, et al. Acute loss of transcription factor E2F1 induces mitochondrial biogenesis in HeLa cells. J Cell Physiol 2006;209:923-34. [PubMed]
- Blanchet E, Annicotte JS, Lagarrigue S, et al. E2F transcription factor-1 regulates oxidative metabolism. Nat Cell Biol 2011;13:1146-52. [PubMed]
- Fang J, Quinones QJ, Holman TL, et al. The H+-linked monocarboxylate transporter (MCT1/SLC16A1): a potential therapeutic target for high-risk neuroblastoma. Mol Pharmacol 2006;70:2108-15. [PubMed]
- Chen SJ, Hoffman NE, Shanmughapriya S, et al. A Splice Variant of the Human Ion Channel TRPM2 Modulates Neuroblastoma Tumor Growth through Hypoxia-inducible Factor (HIF)-1/2α. J Biol Chem 2014;289:36284-302. [PubMed]
- Holmquist-Mengelbier L, Fredlund E, Löfstedt T, et al. Recruitment of HIF-1alpha and HIF-2alpha to common target genes is differentially regulated in neuroblastoma: HIF-2alpha promotes an aggressive phenotype. Cancer Cell 2006;10:413-23. [PubMed]
- Noguera R, Fredlund E, Piqueras M, et al. HIF-1alpha and HIF-2alpha are differentially regulated in vivo in neuroblastoma: high HIF-1alpha correlates negatively to advanced clinical stage and tumor vascularization. Clin Cancer Res 2009;15:7130-6. [PubMed]
- Selak MA, Armour SM, MacKenzie ED, et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-alpha prolyl hydroxylase. Cancer Cell 2005;7:77-85. [PubMed]
- Jögi A, Øra I, Nilsson H, et al. Hypoxia alters gene expression in human neuroblastoma cells toward an immature and neural crest-like phenotype. Proc Natl Acad Sci U S A 2002;99:7021-6. [PubMed]
- Szychot E, Apps J, Pritchard-Jones K. Wilms’ tumor: biology, diagnosis and treatment. Transl Pediatr 2014;3:12-24.
- Metzger ML, Dome JS. Current therapy for Wilms' tumor. Oncologist 2005;10:815-26. [PubMed]
- Pode-Shakked N, Dekel B. Wilms tumor--a renal stem cell malignancy? Pediatr Nephrol 2011;26:1535-43. [PubMed]
- Rakheja D, Chen KS, Liu Y, et al. Somatic mutations in DROSHA and DICER1 impair microRNA biogenesis through distinct mechanisms in Wilms tumours. Nat Commun 2014;2:4802. [PubMed]
- Torrezan GT, Ferreira EN, Nakahata AM, et al. Recurrent somatic mutation in DROSHA induces microRNA profile changes in Wilms tumour. Nat Commun 2014;5:4039. [PubMed]
- Grønskov K, Olsen JH, Sand A, et al. Population-based risk estimates of Wilms tumor in sporadic aniridia. A comprehensive mutation screening procedure of PAX6 identifies 80% of mutations in aniridia. Hum Genet 2001;109:11-8. [PubMed]
- Dome JS, Huff V. Wilms Tumor Overview. In: Pagon RA, Adam MP, Ardinger HH, et al. eds. GeneReviews. Seattle: University of Washington, 1993-2014.
- Davidoff AM. Wilms tumor. Adv Pediatr 2012;59:247-67. [PubMed]
- Drash A, Sherman F, Hartmann WH, et al. A syndrome of pseudohermaphroditism, Wilms' tumor, hypertension, and degenerative renal disease. J Pediatr 1970;76:585-93. [PubMed]
- Weksberg R, Shuman C, Beckwith JB. Beckwith-Wiedemann syndrome. Eur J Hum Genet 2010;18:8-14. [PubMed]
- Baujat G, Rio M, Rossignol S, et al. Paradoxical NSD1 mutations in Beckwith-Wiedemann syndrome and 11p15 anomalies in Sotos syndrome. Am J Hum Genet 2004;74:715-20. [PubMed]
- Alter BP, Rosenberg PS, Brody LC. Clinical and molecular features associated with biallelic mutations in FANCD1/BRCA2. J Med Genet 2007;44:1-9. [PubMed]
- Mathew CG. Fanconi anaemia genes and susceptibility to cancer. Oncogene 2006;25:5875-84. [PubMed]
- Provenzi M, Saettini F, Conter V, et al. Is there a role for FDG-PET for the assessment of treatment efficacy in Wilms' tumor? A case report and literature review. Pediatr Hematol Oncol 2013;30:633-9. [PubMed]
- Jeong YJ, Sohn MH, Lim ST, et al. F-18 FDG PET/CT imaging of metastatic extrarenal Wilms tumor arising in the inguinal canal. Clin Nucl Med 2011;36:475-8. [PubMed]
- Feichtinger RG, Neureiter D, Royer-Pokora B, et al. Heterogeneity of mitochondrial energy metabolism in classical triphasic Wilms' tumor. Front Biosci (Elite Ed) 2011;3:187-93. [PubMed]
- Balsaver AM, Gibley CW Jr, Tessmer CF. Ultrastructural studies in Wilms's tumor. Cancer 1968;22:417-27. [PubMed]
- Hammer E, Ernst FD, Thiele A, et al. Kidney protein profiling of Wilms' tumor patients by analysis of formalin-fixed paraffin-embedded tissue samples. Clin Chim Acta 2014;433:235-41. [PubMed]
- Dungwa JV, Hunt LP, Ramani P. Overexpression of carbonic anhydrase and HIF-1α in Wilms tumours. BMC Cancer 2011;11:390. [PubMed]
- Maturu P, Overwijk WW, Hicks J, et al. Characterization of the inflammatory microenvironment and identification of potential therapeutic targets in wilms tumors. Transl Oncol 2014;7:484-92. [PubMed]
- Marín-Hernández A, Gallardo-Pérez JC, Ralph SJ, et al. HIF-1alpha modulates energy metabolism in cancer cells by inducing over-expression of specific glycolytic isoforms. Mini Rev Med Chem 2009;9:1084-101. [PubMed]
- Skotnicka-Klonowicz G, Kobos J, Łoś E, et al. Prognostic value of p53 expression in Wilms' tumor in children. Med Sci Monit 2001;7:1224-9. [PubMed]
- Bardeesy N, Falkoff D, Petruzzi MJ, et al. Anaplastic Wilms' tumour, a subtype displaying poor prognosis, harbours p53 gene mutations. Nat Genet 1994;7:91-7. [PubMed]
- Rakheja D, Khokhar S, Mitui M, et al. Immunohistochemical expression of GLUT1 and its correlation with unfavorable histology and TP53 codon 72 polymorphism in Wilms tumors. Pediatr Dev Pathol 2012;15:286-92. [PubMed]
- Zawacka-Pankau J, Grinkevich VV, Hünten S, et al. Inhibition of glycolytic enzymes mediated by pharmacologically activated p53: targeting Warburg effect to fight cancer. J Biol Chem 2011;286:41600-15. [PubMed]
- Senanayake U, Das S, Vesely P, et al. miR-192, miR-194, miR-215, miR-200c and miR-141 are downregulated and their common target ACVR2B is strongly expressed in renal childhood neoplasms. Carcinogenesis 2012;33:1014-21. [PubMed]
- Perotti D, Hohenstein P, Bongarzone I, et al. Is Wilms tumor a candidate neoplasia for treatment with WNT/β-catenin pathway modulators?--A report from the renal tumors biology-driven drug development workshop. Mol Cancer Ther 2013;12:2619-27. [PubMed]
- Fukuzawa R, Heathcott RW, Sano M, et al. Myogenesis in Wilms' tumors is associated with mutations of the WT1 gene and activation of Bcl-2 and the Wnt signaling pathway. Pediatr Dev Pathol 2004;7:125-37. [PubMed]
- Maiti S, Alam R, Amos CI, et al. Frequent association of beta-catenin and WT1 mutations in Wilms tumors. Cancer Res 2000;60:6288-92. [PubMed]
- Koesters R, Ridder R, Kopp-Schneider A, et al. Mutational activation of the beta-catenin proto-oncogene is a common event in the development of Wilms' tumors. Cancer Res 1999;59:3880-2. [PubMed]
- Yoon JC, Ng A, Kim BH, et al. Wnt signaling regulates mitochondrial physiology and insulin sensitivity. Genes Dev 2010;24:1507-18. [PubMed]
- Esen E, Chen J, Karner CM, et al. WNT-LRP5 signaling induces Warburg effect through mTORC2 activation during osteoblast differentiation. Cell Metab 2013;17:745-55. [PubMed]
- Da Forno PD, Pringle JH, Hutchinson P, et al. WNT5A expression increases during melanoma progression and correlates with outcome. Clin Cancer Res 2008;14:5825-32. [PubMed]
- Dejmek J, Dejmek A, Säfholm A, et al. Wnt-5a protein expression in primary dukes B colon cancers identifies a subgroup of patients with good prognosis. Cancer Res 2005;65:9142-6. [PubMed]
- Kurayoshi M, Oue N, Yamamoto H, et al. Expression of Wnt-5a is correlated with aggressiveness of gastric cancer by stimulating cell migration and invasion. Cancer Res 2006;66:10439-48. [PubMed]
- Lejeune S, Huguet EL, Hamby A, et al. Wnt5a cloning, expression, and up-regulation in human primary breast cancers. Clin Cancer Res 1995;1:215-22. [PubMed]
- Lee SY, Jeon HM, Ju MK, et al. Wnt/Snail signaling regulates cytochrome C oxidase and glucose metabolism. Cancer Res 2012;72:3607-17. [PubMed]
- Pate KT, Stringari C, Sprowl-Tanio S, et al. Wnt signaling directs a metabolic program of glycolysis and angiogenesis in colon cancer. EMBO J 2014;33:1454-73. [PubMed]
- Nisen PD, Zimmerman KA, Cotter SV, et al. Enhanced expression of the N-myc gene in Wilms' tumors. Cancer Res 1986;46:6217-22. [PubMed]
- Zhang X, Xing G, Saunders GF. Proto-oncogene N-myc promoter is down regulated by the Wilms' tumor suppressor gene WT1. Anticancer Res 1999;19:1641-8. [PubMed]
- Sherwood V. WNT Signaling: an Emerging Mediator of Cancer Cell Metabolism? Mol Cell Biol 2015;35:2-10. [PubMed]
- Maschietto M, Charlton J, Perotti D, et al. The IGF signalling pathway in Wilms tumours--a report from the ENCCA Renal Tumours Biology-driven drug development workshop. Oncotarget 2014;5:8014-26. [PubMed]
- Aiden AP, Rivera MN, Rheinbay E, et al. Wilms tumor chromatin profiles highlight stem cell properties and a renal developmental network. Cell Stem Cell 2010;6:591-602. [PubMed]
- Ruteshouser EC, Robinson SM, Huff V. Wilms tumor genetics: mutations in WT1, WTX, and CTNNB1 account for only about one-third of tumors. Genes Chromosomes Cancer 2008;47:461-70. [PubMed]
- Baudry D, Cabanis MO, Patte C, et al. Cadherins in Wilms' tumor: E-cadherin expression despite absence of WT1. Anticancer Res 2003;23:475-8. [PubMed]
- Xie CH, Naito A, Mizumachi T, et al. Mitochondrial regulation of cancer associated nuclear DNA methylation. Biochem Biophys Res Commun 2007;364:656-61. [PubMed]
- Naito A, Cook CC, Mizumachi T, et al. Progressive tumor features accompany epithelial-mesenchymal transition induced in mitochondrial DNA-depleted cells. Cancer Sci 2008;99:1584-8. [PubMed]
- Little M, Wells C. A clinical overview of WT1 gene mutations. Hum Mutat 1997;9:209-25. [PubMed]
- Schumacher V, Schuhen S, Sonner S, et al. Two molecular subgroups of Wilms' tumors with or without WT1 mutations. Clin Cancer Res 2003;9:2005-14. [PubMed]
- Rae FK, Martinez G, Gillinder KR, et al. Anlaysis of complementary expression profiles following WT1 induction versus repression reveals the cholesterol/fatty acid synthetic pathways as a possible major target of WT1. Oncogene 2004;23:3067-79. [PubMed]
- Niewisch MR, Kuçi Z, Wolburg H, et al. Influence of dichloroacetate (DCA) on lactate production and oxygen consumption in neuroblastoma cells: is DCA a suitable drug for neuroblastoma therapy? Cell Physiol Biochem 2012;29:373-80. [PubMed]
- Vella S, Conti M, Tasso R, et al. Dichloroacetate inhibits neuroblastoma growth by specifically acting against malignant undifferentiated cells. Int J Cancer 2012;130:1484-93. [PubMed]
- Skinner R, Trujillo A, Ma X, et al. Ketone bodies inhibit the viability of human neuroblastoma cells. J Pediatr Surg 2009;44:212-6; discussion 216. [PubMed]
- Rohde BH, Chiou GC. Effects of glucose on neuroblastoma in vitro and in vivo. J Pharm Sci 1987;76:366-70. [PubMed]
- Celay J, Blanco I, Lázcoz P, et al. Changes in gene expression profiling of apoptotic genes in neuroblastoma cell lines upon retinoic acid treatment. PLoS One 2013;8:e62771. [PubMed]
- Knelson EH, Gaviglio AL, Nee JC, et al. Stromal heparan sulfate differentiates neuroblasts to suppress neuroblastoma growth. J Clin Invest 2014;124:3016-31. [PubMed]


