Methylenetetrahydrofolate reductase polymorphisms at 3'-untranslated region are associated with susceptibility to preterm birth
Introduction
Preterm birth [(PTB), delivery before 37 weeks in humans] is the leading cause of neonatal mortality and morbidity, with an incidence between 5-12% (1). In Suzhou, the incidence is 6%. Etiology is complex, including environment and genetic factors involved in both maternal and fetal aspects. Many studies revolved that both maternal and fetal genetic factors contributed to gestational age (2-4). PTB has significant genetic contributions that genetic factors account for 25-40% of variation in gestational duration (5). Recent PTB large-scale genetic studies were published (6). Identifying genetic contributions will help to genetic screening tests and to predict to PTB.
Folic acid metabolism is closely related to many important genetic and epigenetic pathways. It can affect DNA synthesis, amino acid metabolism, methyl donor contents, homocysteine (Hcy) concentration that could give rise to congenital heart disease (CHD) (7) and PTB. Folate deficiency during pregnancy is one of the risk factors for anaemia, low birth weight and PTB. In a cohort of 34,480 singleton pregnancies, preconceptional folate supplementation is associated with a 50-70% reduction in the incidence of spontaneous PTB (8). Growing evidences show that polymorphisms of genes related to folic acid metabolism pathways are involved in PTB. In recent years, several studies focus on association study of single nucleotide polymorphisms (SNPs) with folic acid metabolism and it is reported that SNPs in the coding sequence of several genes are associated with PTB (9-12).
Methylenetetrahydrofolate reductase (MTHFR) is an enzyme mainly involved in folic acid metabolism pathways responsible for the conversion of 5,10-methylenetetrahydrofolate to 5-methyltetrahydrofolate, the predominant form of folate in plasma involved in DNA methylation pathways or purine synthesis (13). Polymorphisms locating in the exons of this gene can result in the transition of amino acid that finally lead to the reduction of enzyme activity. For instance, common polymorphism 677C > T homozygous variant can reduce its activity by 70% compared with wild type (14). Chen et al.’s study shows that the infant MTHFR CT and TT genotypes are responsible for mother’s PTB and low birth weight (15). At present there are few reports of the association of genetic variation at non-coding area including 3'-untranslated region (3'-UTR) of MTHFR with PTB. 3'-UTR, a non-coding region of mRNA, is a target of microRNA (miRNA). miRNAs can regulate the genes translation through sequence-specific binding to 3'-UTR, thus will result in inhibition of translation or degradation of target mRNAs which can reduce the activity of enzyme (16). Recent studies show that polymorphisms in miRNA and 3'-UTR are related with tumors synthesis (17-19) and CHD (7).
We selected three SNPs at 3'-UTR of MTHFR that may disrupt binding capacity of miRNA using miRNA prediction tools (miRnadb, TargetScan and PicTar). Then we performed genotyping of the three SNPs in a case-control study of PTB to identify whether or not they were involved in the susceptibility to PTB.
Materials and methods
Study subjects
A total of 1,135 subjects were enrolled with informed consent, including 480 cases of preterm babies and 655 normal in-term babies who were born in the Suzhou Maternal-child Medical Center from 2012 to 2013. DNA from all samples was extracted from blood spots using NP968 automatic nucleic acid extraction apparatus (Xi’an Tianlong Science and Technology Co. Ltd., China), which was originally used for neonatal screening for inborn errors of metabolism. The study was approved by the Ethic Committee for Human Research, Suzhou Hospital Affiliated to Nanjing Medical University.
SNPs selection
Three prediction tools were used for bioinformatic analysis of the miRNA binding sites at the 3'-UTR of MTHFR: miRnadb, PicTar and TargetScan (20-22). miRNA 22, miRNA24, miR-1304-3p, miR-1224-3p and miR-3150-5p were candidate miRNAs for further study. Nine SNPs at the 3'-UTR of MTHFR including rs145253240 (minor allele frequency; MAF 0.05%), rs45451599 (MAF 0.6%), rs45574135 (MAF unknown), rs186662631 (MAF 0.2%), rs191876689 (MAF <0.01), rs45445997 (MAF 0.2%), rs1537515 (MAF 8.4%), rs1537516 (MAF 8.4%) and rs182398073 (MAF 0.5%) were candidate SNPs. A small cohort of samples including 50 PTB cases and 50 controls were genotyped. Only three SNPs loci, rs1537516, rs1537515 and rs45451599 showed heterozygotic or homozygotic genotypes. So these three SNPs were finally selected for further study.
Genotyping
To understand the potential role of SNPs in genes involved in folate metabolism in PTB, three candidate SNPs in MTHFR were genotyped using the SNaPshot method. The protocol for the SNaPshot method has been published elsewhere and was used with some modifications (23,24). The primers were synthesized by the Shanghai Genearray Company in China. The primers were synthesized by the Genearray Company (Shanghai, China).
Statistics analysis
The χ2 test was performed, using SPSS software (17.0) for Windows. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated when P<0.05 was considered statistically significant.
Results
We screened three candidate SNPs (rs45451599, rs1537515 and rs1537516) at 3'-UTR of MTHFR to determine their association with PTB in 480 PTB babies and 655 term babies using the SNaPshot method. All SNaPShot extension primers were designed in the reverse orientation of NCBI Reference Sequence NC_000001.10. Mutations are shown in B and C marked with an asterisk. There were 281 male and 199 female infants in the PTB group, and 379 male and 276 female infants in the control group. The P value of infants’ gender was 0.842. The genotype frequencies of the three SNPs were in agreement with Hardy-Weinberg equilibrium. Data regarding genotype distributions of three SNPs and χ2 analysis is shown in Table 1. There are mutation frequencies in two SNPs (rs1537515 and rs1537516) showed significant differences between PTB group and control group, 17.9% (117/655) GT heterozygote of rs1537515 were found in the control group, which was significantly higher than that in the PTB group [12.7% (61/480)], suggesting that GT heterozygote of rs1537515 may be a protective factor for PTB (OR: 0.65; 95% CI, 0.46-0.91; and P=0.012). Similarly, CT heterozygote of rs1537516 may be a protective factor for PTB. However, we found genotype distribution of rs45451599 showed no significant differences between the PTB group and the control group, although the AG heterozygote of rs45451599 was 2.6% in the control group, higher than that in the preterm group (1.9%), which suggested that this SNP has no correlation with PTB.
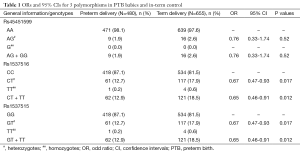
Full table
Furthermore, we also found a consistent protection effect against preterm from the T allele of rs1537516 and rs1537515, compared to the G allele of rs1537515 and C allele of rs1537516 in 34-37 week babies group (per allele OR: 0.677; 95% CI, 0.49-0.94; P=0.021) (Table 2). However, in <32-week group and 32-33-week group, we found no significant correlation between T allele of rs1537516 and rs1537515 with preterm, P>0.05. This contradiction could be explained by the different factors for different gestational age preterm.
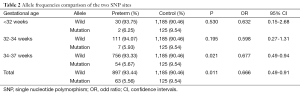
Full table
Discussion
It is reported that preconception supplementation of folic acid can make spontaneous PTB 50-70% reduction in the study of 34,480 pregnant women (8). But its potential pathogenic mechanism is unknown. The molecular epidemiological studies have confirmed that SNPs in folic acid metabolism related genes lead to reduction of the enzyme activity and thus explain the individuals carrying the SNPs with susceptibility to PTB. MTHFR is a key enzyme for intracellular folate homeostasis and metabolism. It is reported that two common MTHFR polymorphisms, MTHFR-677C > T and MTHFR-1298A > C, may be associated with PTB by decreasing the production of the enzyme of MTHFR (12,15). However, these studies focused on SNPs in the coding region and non-coding region have been largely ignored.
It has been proposed that 3'-UTR sequence is recognized via transcriptional and post-transcriptional regulatory pathways. This regulatory region plays an important role in the control of genes expressions. Many recent studies on 3'-UTR function have been reported, and SNPs at 3'-UTR are predicted to play roles in gene expression (25,26) and susceptibility of diseases (27-31), especially in CHD (22). As CHD and PTB shares common folic acid metabolism pathways, it is necessary to research PTB and 3'-UTR. But to our knowledge, there is few articles focusing on the role of MTHFR 3'-UTR in PTB. In that sense, it is necessary to extend from MTHFR coding region to non-coding area. In this article, we analyzed the association between three SNPs locating at 3'-UTR in MTHFR and PTB.
The 3'-UTR of MTHFR has solid predictions for extremely conserved miRNA binding sites, so SNPs in the miRNA target sites may have influence on the gene expression (32). rs1537515 is one of the SNPs in the miRNA target sites, locating in the binding sites of hsa-miR-1224-3p and miR-3150-5p according to software predictions. Based on its special position, rs1537515 attracts us to research its effect on PTB, although there is no study on the association between those miRNAs and MTHFR expression. Firstly, we analyzed genotyping distributions of rs1537515 in Han Chinese population, and the results revealed that the genotyping distributions of rs1537515 were GG 81.5%, GT 17.9% and TT 0.6%. The frequencies of the heterozygous genotype GT of rs1537515 was 12.7% in PTB group, significantly lower than 17.9% in control group. The results indicated that GT of rs1537515 was a protective genetic factor for PTB. As expected, the comparison of allele frequencies showed that wild allele played the leading role. Although, it’s unknown that how to protect from PTB by GT of rs1537515, it gives a new clue to the study of genetic susceptibility to PTB.
Rs1537516 is another SNP in the miRNA target sites, locating at the binding sites of hsa-miR-1304-3p according to prediction. Rs1537516 is 40bp distance from rs1537515. Due to perfectly correlate with rs1537515, it’s proposed that rs1537516 and rs1537515 have the similar regulatory function on MTHFR expression. Indeed, similarly to rs1537515, the frequencies of the heterozygous genotype CT of rs1537516 was 12.7% in PTB group, significantly lower than 17.9% in control group. Our study revealed that CT of rs1537516 was a protective genetic factor for PTB. Same with rs1537516, the comparison of allele frequencies of this site played the leading role in PTB.
Rs45451599 is in miR-24 binding site, however, no significant differences in rs45451599 genotypes were observed among the PTB babies (9/480, 1.9%) and controls (16/655, 2.6%). This may be the result of the small number of AG variants in this case-control study in Han Chinese populations, reducing the power necessary for detectable differences. However, further studies should be performed in different ethnic groups to confirm this result.
Conclusions
In conclusion, this research reveals an association between SNPs at 3'-UTR of MTHFR gene in folate metabolic pathways which may affect the binding capacity of hsa-miR-1304-3p, miR-1224-3p and miR-3150-5p and subsequently increases folate pools and involves in the prevention of PTB. But further functional study should be done for confirmation the significance of these two SNPs in folate metabolic pathways.
Acknowledgements
Funding: This study was partially supported by the “Twelfth Five” Key Medical Talent’s Project in Science and Education of Jiangsu Province (RC2011036); Grant of Natural Science of Jiangsu Province (BK2012600, BK2013095); Jiangsu Provincial Key Discipline Construction Project (FXK201216); Suzhou Science and Technology Development Project (SYSD2011101); Suzhou Key Lab of Translational Medicine Project (SZS201206). The authors are grateful to all the participants for their cooperation in this study.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Goldenberg RL, Culhane JF, Iams JD, et al. Epidemiology and causes of preterm birth. Lancet 2008;371:75-84. [PubMed]
- Boyd HA, Poulsen G, Wohlfahrt J, et al. Maternal contributions to preterm delivery. Am J Epidemiol 2009;170:1358-64. [PubMed]
- York TP, Strauss JF 3rd, Neale MC, et al. Estimating fetal and maternal genetic contributions to premature birth from multiparous pregnancy histories of twins using MCMC and maximum-likelihood approaches. Twin Res Hum Genet 2009;12:333-42. [PubMed]
- Wilcox AJ, Skjaerven R, Lie RT. Familial patterns of preterm delivery: maternal and fetal contributions. Am J Epidemiol 2008;167:474-9. [PubMed]
- Bezold KY, Karjalainen MK, Hallman M, et al. The genomics of preterm birth: from animal models to human studies. Genome Med 2013;5:34. [PubMed]
- McElroy JJ, Gutman CE, Shaffer CM, et al. Maternal coding variants in complement receptor 1 and spontaneous idiopathic preterm birth. Hum Genet 2013;132:935-42. [PubMed]
- Zhao JY, Sun JW, Gu ZY, et al. Genetic polymorphisms of the TYMS gene are not associated with congenital cardiac septal defects in a Han Chinese population. PLoS One 2012;7:e31644. [PubMed]
- Bukowski R, Malone FD, Porter FT, et al. Preconceptional folate supplementation and the risk of spontaneous preterm birth: a cohort study. PLoS Med 2009;6:e1000061. [PubMed]
- Engel SM, Olshan AF, Siega-Riz AM, et al. Polymorphisms in folate metabolizing genes and risk for spontaneous preterm and small-for-gestational age birth. Am J Obstet Gynecol 2006;195:1231.e1-11.
- Johnson WG, Scholl TO, Spychala JR, et al. Common dihydrofolate reductase 19-base pair deletion allele: a novel risk factor for preterm delivery. Am J Clin Nutr 2005;81:664-8. [PubMed]
- Maayan-Metzger A, Lubetsky A, Kuint J, et al. The impact of genetic and environmental factors on homocysteine levels in preterm neonates. Pediatr Blood Cancer 2013;60:659-62. [PubMed]
- Valdez LL, Quintero A, Garcia E, et al. Thrombophilic polymorphisms in preterm delivery. Blood Cells Mol Dis 2004;33:51-6. [PubMed]
- Ryan BM, Weir DG. Relevance of folate metabolism in the pathogenesis of colorectal cancer. J Lab Clin Med 2001;138:164-76. [PubMed]
- Ulrich CM, Robien K, Sparks R. Pharmacogenetics and folate metabolism —a promising direction. Pharmacogenomics 2002;3:299-313. [PubMed]
- Chen DF, Hu YH, Yang F, et al. Mother's and child's methylenetetrahydrofolate reductase C677T polymorphism is associated with preterm delivery and low birth weight. Beijing Da Xue Xue Bao 2004;36:248-53. [PubMed]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116:281-97. [PubMed]
- Glinsky GV. Disease phenocode analysis identifies SNP-guided microRNA maps (MirMaps) associated with human "master" disease genes. Cell Cycle 2008;7:3680-94. [PubMed]
- Landi D, Gemignani F, Barale R, et al. A catalog of polymorphisms falling in microRNA-binding regions of cancer genes. DNA Cell Biol 2008;27:35-43. [PubMed]
- Yu Z, Li Z, Jolicoeur N, et al. Aberrant allele frequencies of the SNPs located in microRNA target sites are potentially associated with human cancers. Nucleic Acids Res 2007;35:4535-41. [PubMed]
- Martin MM, Buckenberger JA, Jiang J, et al. The human angiotensin II type 1 receptor +1166 A/C polymorphism attenuates microRNA-155 binding. J Biol Chem 2007;282:24262-9. [PubMed]
- Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer 2010;10:389-402. [PubMed]
- Wu C, Gong Y, Sun A, et al. The human MTHFR rs4846049 polymorphism increases coronary heart disease risk through modifying miRNA binding. Nutr Metab Cardiovasc Dis 2013;23:693-8. [PubMed]
- Bardien S, Human H, Harris T, et al. A rapid method for detection of five known mutations associated with aminoglycoside-induced deafness. BMC Med Genet 2009;10:2. [PubMed]
- Wu CC, Lu YC, Chen PJ, et al. Application of SNaPshot multiplex assays for simultaneous multigene mutation screening in patients with idiopathic sensorineural hearing impairment. Laryngoscope 2009;119:2411-6. [PubMed]
- Hino K, Sato H, Sugai A, et al. Downregulation of Nipah virus N mRNA occurs through interaction between its 3' untranslated region and hnRNP D. J Virol 2013;87:6582-8. [PubMed]
- Melanson BD, Cabrita MA, Bose R, et al. A novel cis-acting element from the 3'UTR of DNA damage-binding protein 2 mRNA links transcriptional and post-transcriptional regulation of gene expression. Nucleic Acids Res 2013;41:5692-703. [PubMed]
- Adams LA, Möller M, Nebel A, et al. Polymorphisms in MC3R promoter and CTSZ 3'UTR are associated with tuberculosis susceptibility. Eur J Hum Genet 2011;19:676-81. [PubMed]
- Akhter Q, Masood A, Ashraf R, et al. Polymorphisms in the 3'UTR of the human leptin gene and their role in hypertension. Mol Med Rep 2012;5:1058-62. [PubMed]
- Chen Z, Brant SR, Li C, et al. CTLA4 -1661A/G and 3'UTR long repeat polymorphisms are associated with ulcerative colitis and influence CTLA4 mRNA and protein expression. Genes Immun 2010;11:573-83. [PubMed]
- Pan ZW, Luo CF, Liu ZJ, et al. RET 3'UTR polymorphisms and its protective role in Hirschsprung disease in southeastern Chinese. J Pediatr Surg 2012;47:1699-705. [PubMed]
- Silva ID, Muniz YC, Sousa MC, et al. HLA-G 3'UTR polymorphisms in high grade and invasive cervico-vaginal cancer. Hum Immunol 2013;74:452-8. [PubMed]
- Stone N, Pangilinan F, Molloy AM, et al. Bioinformatic and genetic association analysis of microRNA target sites in one-carbon metabolism genes. PLoS One 2011;6:e21851. [PubMed]


