Cloning and sequence analysis of SLC25A13 transcripts in human amniocytes
Introduction
Neonatal Intrahepatic Cholestasis caused by Citrin Deficiency (NICCD, OMIM #605814) is an autosomal recessive inherited disease due to mutations of SLC25A13 gene (1,2). The pathogenic age of NICCD centers in neonate and infancy, and the main manifestations are hepatomegaly, jaundice, and abnormal findings at liver function analysis. Meanwhile, NICCD is now recognized as a world-wide distribution disease (1-13). NICCD is not rare in China, and it was proved that the rate of SLC25A13 mutation carriers was as high as 1/48 in the South China by molecular epidemiological investigation, the frequency of homozygote was then calculated to be 1/9,200 (4,5). Through high-risk screening for inborn errors of metabolism (IEMs) by urease pretreatment-gas chromatography-mass spectrometry (UP-GC-MS), the positive rate of NICCD was ranked the second in Chinese IEM spectrum (8). Meanwhile, a single-center clinical study suggested that NICCD was a major cause for cholestatic liver disease in Chinese children (9).
More and more clinical practice at home and abroad showed that part of NICCD patients would present as a new clinical phenotype: Failure to Thrive and Dyslipidemia caused by Citrin Deficiency (FTTDCD) (11,14), part of them would pass away due to fatal liver cirrhosis or severe complications including intracranial infection (10,11,15,16), and some had to receive liver transplantation to survive (17,18). So the health hazards of NICCD to children can not be ignored. It is obvious that early diagnosis, including prenatal diagnosis, is of great significance and should gain more concern in the medical profession.
For now, the most reliable diagnostic basis for NICCD is gene analysis including screening the high frequency of SLC25A13 mutations by PCR/LA-PCR gel electrophoresis, PCR restriction fragment length polymorphism (RFLP), and identifying the novel mutations by sequencing analysis of DNA fragments amplified by PCR or LA-PCR. However, these diagnosis technologies were mainly at genomic DNA level. Although there were a few reports on the mutations analysis at the mRNA level, these studies were limited to fragment but not the full-length cDNA encoding sequence (1,12,13). It has been reported that about 15% of mutation types can not be accurately determined with the genomic analysis (5). Furthermore, how to analyze the function of proteins encoded by SLC25A13 mutation reliably and fastly is still an unresolved issue (i.e. the completeness and accuracy of amino acid sequence of protein can not be determined by Western blot) (19). In view of the current research status of NICCD, we studied the transcription of the SLC25A13 gene in cultured amniocytes, and this study will provide the basis for prenatal diagnosis of NICCD at the mRNA level.
Materials and methods
Reagents
Genomic DNA extraction kit (Simgen); DNA Marker, Taq and LA-Taq polymerase (TaKaRa); Gel Extraction Kit (Qiagen); TRIzol® RNA Isolation Reagents (Ambion); M-MLV Reverse Transcriptase (Promega); Primer synthesis (Invitrogen).
Origin of human amniocytes
In this paper, two cultured adherent amniocytes were taken as research subjects. One sample was collected from a pregnant woman with amniocentesis for the purpose of diagnosis of Citrin deficiency. The fetus in this sample, whose parents was both 851del4 carriers and previously had a child who suffered from Citrin deficiency (851del4/851del4) and died at 13.5 months of age due to liver failure, was proved to be an 851del4 mutation carrier by genomic DNA analysis (16). The other amniocyte sample, as a control, was from a pregnant woman whose fetus had a high risk of chromosomal aberration so that she was asked to carry out prenatal diagnosis, whereas there was no familial history of Citrin deficiency in this family. The protocol was approved by Committee of Medical Ethics in The First Affiliated Hospital of Jinan University, with informed consents from all two families.
Mutation detection in SLC25A13 exons and their flanking sequences
The genomic DNA of amniocytes was extracted with genomic DNA extraction kit (Simgen) in accordance with instructions. For mutation detection in SLC25A13 exons and their flanking sequences, we mainly used sequencing analysis of DNA fragments amplified by genomic DNA-PCR, as described previously (10,12).
ORF amplification of SLC25A13 cDNA by nest PCR
Primer design
Three primers (RAS2, RACEA1 and RAS3) were designed based on human SLC25A13 mRNA sequence (GenBank ID: AF118838), and the primer Ex18R was cited from the literature (2). Primer RAS2 (5′- AACGCACGCTGCCTGGCCGTATC-3′) and RACEA1 (5′-CCACCTTCACAAATTCATGCGCC-3′) were used in the first PCR, and the deduced amplicon size was 3107 bp; Primer RAS3 (5′-GCCGCCGGGACTAGAAGTGAGC-3′) and Ex18R (5′-TGCTTCATTCCCAGGAGGGA-3′) were used in the second PCR, and the deduced amplicon size was 2191 bp, which contained the entire coding portion of SLC25A13 cDNA.
RNA extraction and cDNA synthesis
Total RNA was extracted from cultured amniocytes with RNAiso Plus (TaKaRa) according to the manufacture’s protocol. The concentration and quality of total RNA were verified with spectrophotometer (Bio-Rad) and electrophoresis with 1.2% agarose gel. Oligo-(dT)18 primed cDNAs were synthesized from 2 µg of total RNA (OD260/OD280=1.8-2.0) in the presence of 1 µg oligo-(dT)18 and 200 U M-MLV reverse transcriptase (Promega) following the manufacturer’s protocol.
Nest PCR
The first PCR was carried out in a 50 µL mixture containing 10 µL of 5 × PrimeSTAR® Buffer (Mg2+ plus) (TaKaRa), 4 µL of dNTP (2.5 mM), 1µL of each primer RAS2 and RACEA1 (20 µM), 31.5 µL of PCR-grade water, 0.5 µL of PrimeSTAR® HS DNA Polymerase (2.5 U/µL, TaKaRa) and 2 µL of cDNA. After initial denature at 94 °C for 3 min, 20 cycles of amplification were performed (98 °C for 10 s, 60 °C for 15 s, 72 °C for 4 min), followed by terminal extension at 72 °C for 7 min.
Subsequently, 1 µL of first PCR products was subjected to the second PCR for 30 cycles in 50 µL mixture, containing 10 µL of 5 × PrimeSTAR® Buffer (Mg2+ plus) (TaKaRa), 4 µL of dNTP (2.5 mM), 1 µL of each primer RAS3 and Ex18R (20 µM), 32.5 µL of PCR-grade water, 0.5 µL of PrimeSTAR® HS DNA Polymerase (2.5 U/µL, TaKaRa). The PCR temperature profile was 94 °C for 3 min followed by 30 cycles of 98 °C for 10 s, 60 °C for 5 s, 72 °C for 2.5 min and a final extension step at 72 °C for 10 min. The amplicon of about 2,200 bp was purified, cloned and sequenced as described below.
Purification, Cloning and sequencing of PCR product
PCR products were separated by electrophoresis with 1.5% agarose gel. The fragments of interest were excised and then purified by gel extraction kit (Qiagen). The purified fragments were linked into pSIMPLE-18 EcoR V/BAP Vector (TaKaRa) by using DNA Kination Kit (D402, TaKaRa) and then transfected into DH5α Escherichia coli cells. The positive clones were identified by colony PCR with a pair of primers RAS3 and Ex18R, and then sequenced. The sequencing results were aligned with the genomic DNA and mRNA sequence of SLC25A13 to analyze the alternative splicing pattern with DNAman Software.
Results
Genomic DNA sequencing results of two cultured amniocytes
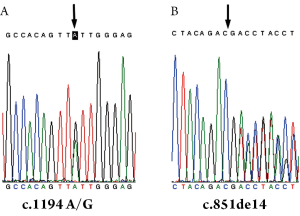
The Genomic DNA sequencing results of the control sample confirmed that there was a SNP in the 17th nt of exon 12 (A/G) (Figure 1A) but no mutations in SLC25A13 gene. However, the results of the other sample showed that the fetus was a carrier of 851del4 mutation (Figure 1B), the same phenotype as his parents.
Results of nest PCR to amplify the ORF of SLC25A13 cDNA
In two samples, there were no clear bands in the first PCR amplification. However, an about 2.2 kb long product was amplified in the second PCR (Figure 2).

Cloning of entire ORF of SLC25A13 cDNA and identification of alternative splicing transcripts
Seven to eight positive clones were picked randomly from each sample and sequenced, respectively, and the analysis results were shown in Figure 3. From the total seven clones of the control sample, we found that, one contained CAG insertion while the others were normal. The 17th base of exon 12 was A in three transcripts but G in the other four transcripts. It demonstrated that two alleles (one inherited from each parent) were both transcribed, so the amniocyte could be used as material to research SLC25A13 transcription. From the total eight clones of 851del4 carrier sample, one exon 5-11 skipping transcript and 7 normal transcripts, but no GTAT deletion, were identified. These results indicated that the SLC25A13 transcripts from the allele with 851del4 mutation were unstable.

Discussion
Point mutations or short deletion/insertion in the exons and their flanking sequences (the main area resulting in abnormal splicing) of SLC25A13 gene could be revealed with genomic DNA analysis method (1,12). It couldn’t be convenient and rapid to get the whole sequence information of SLC25A13 gene since total length of the gene was about 200 kb. However, cDNA sequence analysis could overcome this disadvantage in part. By cDNA cloning and sequence analysis, we can investigate whether or not the two alleles both express normally, analyze sequence information of the entire ORF, and identify abnormal splicing transcripts, so as to find valuable clues to identify novel mutations. Moreover, combined with genomic DNA analysis, SLC25A13 transcripts analysis in amniocytes could improve the reliability of prenatal diagnosis.
At amniocentesis, it was difficult to avoid maternal blood contamination, and impractical to perform sampling repeatedly (20). Once the sample was contaminated with maternal blood, even trace amount, the analysis of PCR-amplified genomic fragments will be difficult, even resulting in misdiagnosis (21). Fetal amniocytes could adhere to the wall of the culture flask, whereas blood cells could not. To avoid maternal blood contamination, amniocytes were cultured in situ and only adherent cells were used in this study. Nest PCR could significantly improve the specificity and sensitivity of DNA amplification. However, the possibility of errors due to wrong amplification also increased. Therefore, contamination should be reduced as far as possible in the process of mRNA extraction, reverse transcription and PCR, and control group should be set up strictly. Human SLC25A13 is mainly expressed in hepatocytes (1), but so far, it has not been reported whether the gene is transcribed in amniocytes. Nest PCR method was used to ensure a successful amplification of the target gene in this study. As showed in Figure 2, the size of the amplified band was appropriate, and sequencing results also demonstrated that the band contained citrin coding region of SLC25A13. This finding indicated that the full length of the coding region in SLC25A13 gene was amplified successfully by nest PCR, providing scientific evidence for prenatal diagnosis of citrin deficiency with amniocytes mRNA analysis.
The transcript with CAG insertion has been included in Ensemble database (ENST00000416240). However, the exon 5-11 skipping transcript was a novel one. It was reported recently that 92-94% of human multi-exon genes undergo alternative splicing, and different transcripts played different roles under strict supervision in different tissues. For example, two alternatively spliced forms varying in C-terminal properties of NAD+-dependent isocitrate dehydrogenase (IDH) possessed with tissue-specific expression, suggesting that alternative splicing was a regulatory mechanism along specific tissue (22,23). Meanwhile, the incidence of alternative splicing was associated closely with development of diseases such as inflammation pathways and cancer (24,25). The significance of the novel SLC25A13 alternative splicing transcripts identified in this paper needs to be further explored.
Due to the limitation of medical ethic and sample sources, only two amniocytes were analyzed in this study. It was found that both SLC25A13 alleles were expressed normally in the amniocytes without Citrin deficiency. However, no transcript containing GTAT deletion was detected in all the 8 clones from amniocytes of the 851del4 carring fetus. This might be the result of nonsense-mediated mRNA decay (NMD), which selectively degrades mRNA that prematurely terminate translation because of frameshift or nonsense mutation, as a conserved mechanism in mRNA surveillance commonly existing in eukaryotic cells (26,27). The 851del4 mutation in SLC25A13 gene led to frameshift after deletion site, produced early stop codon (1), and then initiated NMD. So no transcripts containing 851del4 deletion could be detected.
Acknowledgements
Funding: This research got financial support from the project 81070279 supported by the National Natural Science Foundation of China (NSFC).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Kobayashi K, Sinasac DS, Iijima M, et al. The gene mutated in adult-onset type II citrullinaemia encodes a putative mitochondrial carrier protein. Nat Genet 1999;22:159-63. [PubMed]
- Saheki T, Kobayashi K. Mitochondrial aspartate glutamate carrier (citrin) deficiency as the cause of adult-onset type II citrullinemia (CTLN2) and idiopathic neonatal hepatitis (NICCD). J Hum Genet 2002;47:333-41. [PubMed]
- Dimmock D, Maranda B, Dionisi-Vici C, et al. Citrin deficiency, a perplexing global disorder. Mol Genet Metab 2009;96:44-9. [PubMed]
- Kobayashi K, Ushikai M, Song YZ, et al. Overview of citrin deficiency: SLC25A13 mutations and the frequency. J Appl Clin Pediatr 2008;23:1553-7.
- Lu YB, Kobayashi K, Ushikai M, et al. Frequency and distribution in East Asia of 12 mutations identified in the SLC25A13 gene of Japanese patients with citrin deficiency. J Hum Genet 2005;50:338-46. [PubMed]
- Song YZ, Kobayashi K. Citrin Dificency. J Appl Clin Pediatr 2008;23:1564-5.
- Song YZ, Hao H, Ushikai M, et al. A difficult and complicated case study: neonatal intrahepatic cholestasis caused by citrin deficiency. Zhongguo Dang Dai Er Ke Za Zhi 2006;8:125-8. [PubMed]
- Song YZ, Li BX, Hao H, et al. Selective screening for inborn errors of metabolism and secondary methylmalonic aciduria in pregnancy at high risk district of neural tube defects: a human metabolome study by GC-MS in China. Clin Biochem 2008;41:616-20. [PubMed]
- Song YZ, Ushikai M, Kobayashi K, et al. Citrin deficiency is an important etiology for cholestatic liver disease in children. Zhonghua Er Ke Za Zhi 2009;47:624-7. [PubMed]
- Song YZ, Li BX, Chen FP, et al. Neonatal intrahepatic cholestasis caused by citrin deficiency: clinical and laboratory investigation of 13 subjects in mainland of China. Dig Liver Dis 2009;41:683-9. [PubMed]
- Song YZ, Deng M, Chen FP, et al. Genotypic and phenotypic features of citrin deficiency: five-year experience in a Chinese pediatric center. Int J Mol Med 2011;28:33-40. [PubMed]
- Tabata A, Sheng JS, Ushikai M, et al. Identification of 13 novel mutations including a retrotransposal insertion in SLC25A13 gene and frequency of 30 mutations found in patients with citrin deficiency. J Hum Genet 2008;53:534-45. [PubMed]
- Wen PQ, Wang GB, Chen ZL, et al. SLC25A13 gene analysis in neonates with intrahepatic cholestasis caused by citrin deficiency. Zhongguo Dang Dai Er Ke Za Zhi 2011;13:303-8. [PubMed]
- Song YZ, Guo L, Yang YL, et al. Failure to thrive and dyslipidemia caused by citrin deficiency: a novel clinical phenotype. Zhongguo Dang Dai Er Ke Za Zhi 2009;11:328-32. [PubMed]
- Xing YZ, Qiu WJ, Ye J, et al. Studies on the clinical manifestation and SLC25A13 gene mutation of Chinese patients with neonatal intrahepatic cholestasis caused by citrin deficiency. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2010;27:180-5. [PubMed]
- Zhao XJ, Tang XM, Zha QB, et al. Prenatal diagnosis of citrin deficiency in a Chinese family with a fatal proband. Tohoku J Exp Med 2011;225:273-6. [PubMed]
- Ohura T, Kobayashi K, Tazawa Y, et al. Clinical pictures of 75 patients with neonatal intrahepatic cholestasis caused by citrin deficiency (NICCD). J Inherit Metab Dis 2007;30:139-44. [PubMed]
- Shigeta T, Kasahara M, Kimura T, et al. Liver transplantation for an infant with neonatal intrahepatic cholestasis caused by citrin deficiency using heterozygote living donor. Pediatr Transplant 2010;14:E86-8. [PubMed]
- Tokuhara D, Iijima M, Tamamori A, et al. Novel diagnostic approach to citrin deficiency: analysis of citrin protein in lymphocytes. Mol Genet Metab 2007;90:30-6. [PubMed]
- Chen JH, Lin JY. Application of amniocentesis in prenatal diagnosis of thalassemia. International Medicine and Health Guidance News 2009;15:44-6.
- Huang Y, Li DM. The application of improved amniotic fluid cells culture method in situ in prenatal diagnosis. Guangxi Medical Journal 2008;30:1852-3.
- Wang ET, Sandberg R, Luo S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature 2008;456:470-6. [PubMed]
- Weiss C, Zeng Y, Huang J, et al. Bovine NAD+-dependent isocitrate dehydrogenase: alternative splicing and tissue-dependent expression of subunit 1. Biochemistry 2000;39:1807-16. [PubMed]
- Häsler R, Kerick M, Mah N, et al. Alterations of pre-mRNA splicing in human inflammatory bowel disease. Eur J Cell Biol 2011;90:603-11. [PubMed]
- Hansson O, Zhou Y, Renström E, et al. Molecular function of TCF7L2: Consequences of TCF7L2 splicing for molecular function and risk for type 2 diabetes. Curr Diab Rep 2010;10:444-51. [PubMed]
- Maquat LE. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 2004;5:89-99. [PubMed]
- Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet 1999;8:1893-900. [PubMed]


