Novel deletion and 2397 G>T mutations of the EXT1 gene identified in two Chinese pedigrees with hereditary multiple exostoses using exon sequencing
Introduction
Hereditary multiple exostoses (HME), an autosomal dominant genetic disease, is characterized by chondrogenic osteogenesis and multiple osteochondroma. In Western countries, HME has a prevalence of 1/50,000 (1,2). HME clinically manifests as cartilage-capped bony protrusions from the long metaphyseal end perpendicular to the long bone. Bone warts can occur in any bone, but long bones are especially susceptible and the distal end of the femur and the upper end of the tibia are sites with a high probability of occurrence. Other clinical manifestations include chronic pain syndromes, limb dysfunction, skeletal developmental deformity, short stature, scoliosis, and neurovascular alteration. Although HME is a benign disease, 1–2% of patients progress to chondrosarcoma, a malignant tumor (3). Currently, the drug treatment for multiple osteochondroma is still in the laboratory research phase. Some researchers screen small molecules that function in a similar manner within chondrocyte cultures, as well as, in reducing the development and frequency of osteochondromas within various mouse models of HME. The current treatment method of HME is still based on surgery. Surgery is the recommended treatment for removing tumors and improving limb function and the appearance of deformities, as well as for suspected cases of malignant transformation. Simple tumor resection is mainly suitable for large tumors and tumors that compress the surrounding muscle and nerves (4,5). However, the best surgical timing and method for patients with different disease stages and needs have proved controversial. Previous studies have discussed several theories on the pathogenesis of HME such as abnormal embryo growth and development, dislocation and displacement of growth plate and the metaphyseal periosteum is not fully developed. However, its exact pathogenic mechanism is still unclear. The mechanism of HME is an extremely complicated process. Because of its genetic heterogeneity, HME and mutations of EXT (exotosin) are interrelated in molecular genetics research. Most of the pathogenic genes found in studies to date have been concentrated in the EXT1 (exotosin-1, MIM # 608117) gene and the EXT2 (exotosin-2, MIM # 608210) gene (6). In previous studies, frameshift, nonsense, or splicing mutations have been the most common types of EXT mutations found, and large fragment mutations have rarely emerged (7). Mutations in the EXT1 or EXT2 gene lead to partial loss of the function of its encoded protein, which lead to cartilage internalization, bone development disorders and osteochondroma formation which can be through signal transduction systems such as Ihh, FGF, and BMP.
The EXT1 gene located on chromosome 8q23-q24 and the EXT2 gene located on chromosome 11p11-p12 contain 11 and 16 exons, respectively. Linkage analysis identified the EXT3 gene on chromosome 19p (8). However, the EXT3 gene has not yet been cloned and related research is limited. EXT1 and EXT2 mutations were found in 70–94% of patients with human HME and can exist in one or both genes. EXT1 and EXT2 genes are highly homologous (9). Their coding products localized in the Golgi apparatus are widely present in all somatic cells as type II transmembrane glycoproteins (10,11). The two form a heterologous oligomeric complex, heparan sulfate polymerase, which has a high level of glycosyltransferase activity. Heparan sulfate polymerase, a heparan sulfate synthetase, catalyzes the alternate attachment of glucuronic acid and N-acetyl glucosamine to extend the peptide chain, forming heparan sulfate. The heparan sulfate then covalently bonds to the core protein to form heparan sulfate proteoglycan (HSPG). Proteoglycans exist on the surface of chondrocyte membranes and extracellular matrix involved in signal transduction, cell proliferation, and differentiation (12,13). The germline EXT mutation can influence the production of proteoglycans and the proliferation and differentiation of chondrocytes, which further reduces HS production at perichondrial sites, thus facilitating the formation of osteochondroma. In our previous research, we found three updated mutations of EXT1 (p.Thr332Ser, p.Arg405Lys, and p.Trp412Arg) which are important to the prevention and diagnosis of HME. Despite the recent significant advances, the study of the pathogenic mechanism of HME is still underway, and methods for treatment have yet to be developed. HME still plagues the quality of life of most patients. Given the relationship between the EXT gene and HME, novel insights into the specific role of EXT1 in HME are still greatly needed. The present study focuses on the novel mutations of EXT1 that has a close contact with the occurrence of HME. The quality of life and health complications for participants in our study are exactly coincided with HME, which may own great research value. The research with latest technology aims to determine related mutations that have not been identified before. We present the following article in accordance with the MDAR reporting checklist (available at http://dx.doi.org/10.21037/tp-20-191).
Methods
Pedigree survey and sample collection
Two families were recruited from the Department of Orthopedics of Shanghai Children’s Hospital between June, 2017 and November, 2018. Venous blood samples were collected from the study participants. Genomic DNA was extracted from peripheral blood according to the standard procedures for exome sequencing. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of the Department of Orthopedics of Shanghai Children’s Hospital (NO.: 2017023) and informed consent was taken from all the patients.
Extraction of DNA and RNA
Venous blood samples were collected in EDTA-anticoagulant vacuum blood collection tubes from 11 pedigrees and stored in −20 °C for the research. Total DNA and RNA were extracted from the blood according to the manufacturer’s instructions. The concentration and purity of the DNA and RNA were detected by measuring the absorbance at 260 and at 280 nm with the DU800 UV spectrophotometer (Beckman, USA).
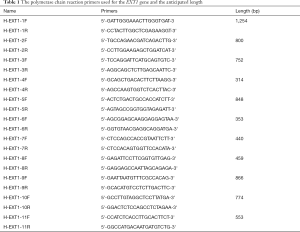
Sequencing of the EXT1 gene and design of primers
The DNA and mRNA sequences of the EXT1 gene (GenBank NG_007455; NM_000127) to verify the exon regions were obtained from the NCBI website (www.ncbi.nlm.nih.gov/). According to the coding region of the EXT gene, the exon and intron junction regions, primers were designed using software, including 11 pairs of EXT1 gene polymerase chain reaction (PCR) primers and 5 pairs of cross-intron primers to cover the 11 coding regions of the EXT1 gene and their flanking sequences. The proportion of GC was 50–55% in these primers and the annealing temperature was 57 °C. The primers were synthesized by Shanghai Jierui Biotech Biotech Co., Ltd. (Tables 1,2). PCR for the EXT1 gene was performed with the DNA templates of the family members using a kit with GC Buffer (Takara, Japan). The PCR protocol was consistent with our previous method. The mixture used for PCR (25 µL) included 3 µL of template, 1 µL of each primer, 2 µL of 2.5 mM dNTP, 0.2 µL of EX TaqE, 12.5 µL of 2× GC buffer and 5.3 µL of ddH2O. All PCR programs conditions can be listed as follows: pre-denaturation at 95 °C for 5 min, a total of 32 cycles of denaturation at 95 °C for 45 s, annealing at 57 °C for 45 s and extension at 72 °C for 45 s, and a final extension at 72 °C for 10 min. The PCR products were stored at 4 °C. When the concentration and purity of the PCR products were up to standard, they were subjected to sequencing by the Shanghai Majorbio Company. The purified PCR products sequenced with BigDye 3.1 (Applied Biosystems, life technologies) in the following PCR programme: denaturation 4 min at 94 °C, 25 cycles of 30 sec at 94 °C, 20 sec at 50 °C and 3 min at 60 °C. The sequences were analyzed with an ABI PRISM 3730 DNA Analyzer (Applied Biosystems, Thermo Fisher Scientific, Inc.). All the mutations detected in the period was confirmed by digestion of each PCR product with the corresponding enzyme and experienced strict quality control. The possible pathogenic mutations screened from the sequencing were manually re-analyzed and would be proved to be novel variants.
Full table

Full table
Detection of DNA and RNA amplification primers
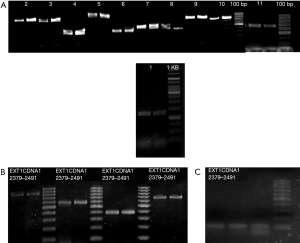
The results of PCR amplification of all exons of the EXT1 gene after agarose gel electrophoresis and imaging are shown in Figure 1. All of the specific primer amplification bands were clear. The length of the amplified fragment was about 100–1,200 bp, which is in line with the target PCR amplification, and the PCR product could be sequenced.
RT-qPCR
Total RNA was extracted from the venous blood samples using the methods previously described. cDNA was generated with a reverse transcription kit (Promega, Madison, Wisconsin, USA). Quantitative PCR reagents were applied to detect the mRNA level of EXT1. Real-time PCR was performed according to manufacturer’s protocol.
Statistical analysis
All data were expressed as the mean ± SEM. Comparisons between two groups were made using Student’s t-test. A P value of <0.05 was considered to be statistically significant. Statistical analyses were performed with GraphPad Prism 6.0 (Graph Pad Prism Software) and IBM SPSS Statistics version 14.0 for Windows (IBM Corp, New York, USA).
Results
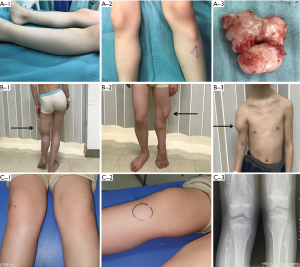
Clinical manifestation
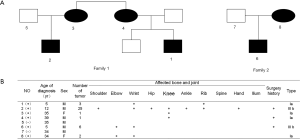
The baseline characteristics of the two families are presented in Figure 2. All of the participants underwent a general examination and were classified according to the clinical criteria of HME. The results showed that the males were more likely to suffer from HME than the females and present with more severe symptoms. The tumors were located at multiple joints, but most commonly the wrist and knee joints. Spine and iliac joint involvement had not yet occurred, and the patients whose conditions were more severe were younger than the others. The Clinical phenotype of the HME patients was presented in Figure S1.
Sequencing of the EXT1 gene
To reveal the genetic determinant of HME in the participants, each exon of the EXT1 gene was sequenced. According to the sequencing results on NCBI, the mutation sites on the exon of the EXT1 gene and the amino acid changes were obtained by BLAST alignment and triple codon analysis, respectively (Table 3). The results showed that there were mutations in 4 exons of the EXT1 gene: c.1838 C>T in exon 3; c.2534 G>A in exon 9; c.2702 G>T in exon 10; c. absence of exon 7; and 2397 G>T in exon7. The mutations included synonymous, deletion, and nonsense mutations; however, there were no frameshift or large-fragment repeated mutations found. Through codon analysis, it was concluded that mutations in the bases of the corresponding exons can cause changes in single nucleotides as follows: the 1838 C → T mutation in exon 3 corresponds to cysteine → cysteine at 355; the G → A mutation at position 2534 in exon 9 corresponds to valley 587 Glutamate → Glutamate; the 2702 G → T mutation in exon 10 corresponds to lysine → lysine at position 643; and the 2397 G → T mutation in exon 7 corresponds to Glutamate to a stop codon at position 542. Also, the deletion of exon 7 corresponds to the deletion of 32 amino acids. Among these mutations, the deletion of exon 7 and the 2397 G → T mutation on exon 7 have not been reported in previous studies. The presence of the mutation on exon 7 was found in the affected family members but not in the unaffected individuals. Therefore, we may conclude that the novel mutations in EXT1, the absence of exon 7 and 2397 G>T in exon 7, were the molecular and pathophysiological mechanisms underlying HME in patients 1, 2, 3, 4, 6, and 8.

Full table
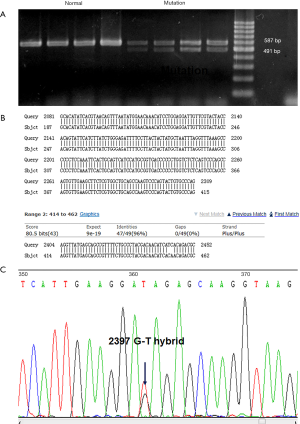
The validation of mutant gene by Sanger sequencing
The results of exon sequencing of EXT1 showed that the deletion of exon 7 and 2397 G>T in exon 7 may exert an impact on gene function. Gene amplification was performed to identify the change of gene expression caused by the mutations. The amplified bands of normal (2310–2405) and mutant samples of exon 7 were 587 and 491 bp, respectively (Figure 3A). The mutant samples were confirmed to be heterozygous for deletion of exon 7. Sanger sequencing was performed to verify the mutation in the mutant samples, and we found that deletion of exon 7 and G>T mutation at base 2397 caused a nonsense mutation at amino acid 542 (Figure 3B,C). As the unaffected relatives did not carry the mutation, deletion of exon 7 and 2397 G>T in exon 7 were seen as candidate pathogenic gene mutations.
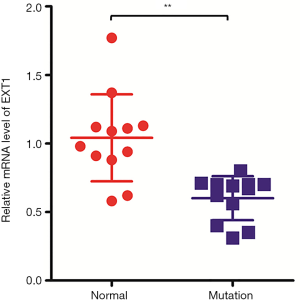
mRNA expression of EXT1 in mutant samples
To define the influence of the deletion mutation and nonsense mutation on EXT1 function, we detected mRNA expression of the EXT1 gene by RT-PCR. The results showed that the relative mRNA expression of the EXT1 gene in normal samples was 1.04±0.31, and the relative expression of the EXT1 gene in samples with exon 7 deletion and 2397 G>T mutation was 0.6±0.14. The mRNA expression of EXT1 decreased by 40%, and the P value was <0.01 with a statistically significant difference (Figure 4), which indicated that deletion of exon 7 and 2397 G>T in exon 7 could greatly impact the expression and function of EXT1.
Discussion
The specific pathological and biochemical mechanisms of HME, an autosomal dominant inheritance cartilage dysplasia, have yet to be fully illuminated. Previous studies have found that the occurrence of HME can be triggered by a variety of factors, including environmental and genetic factors. In particular, genetic mutations play an important role. Our study recruited two families including HME patients and their relatives. These participants were sequenced for the EXT1 gene and the changes of 11 exons in EXT1 were observed. The deletion of exon 7 and the change of base G to T at position 2397 in exon 7 have not been reported previously. The mutations on exon 7 truncate mRNA expression of EXT1, which causes abnormal HS synthesis and leads to abnormal chondrocyte differentiation, eventually triggering a series of HME symptoms.
The most common mutations identified in HME patients are frameshift mutations, followed by nonsense mutations. Deletions, especially the deletion of single exon, are rare. Previous studies have shown that the mutations in EXT1 exon are mainly concentrated in four exons (1, 2, 6, and 8), while the sequences of the remaining exons of the EXT1 gene are conservative, which means the mutations do not easily occur. The results of our study revealed that mutations were present in four exons of EXT1 (exon 3, exon 7, exon 9, and exon 10), most of which were synonymous mutations apart from the deletion mutation and nonsense mutation in exon 7. In recent years, some studies have used exon sequencing or transcriptomics technologies to investigate deletion and nonsense mutations in EXT1 (14,15), but there have been no studies specifically about exon 7. Exon 7 is 96 bp long and codes for 32 amino acids. Its deletion, which rarely occurs, results in the absence of 32 amino acids. Moreover, the 2397 G>T mutation in exon 7 corresponds with the occurrence of glutamate, which is a stop codon that shortens the mRNA expression of EXT1. The above mutations could exert an influence on the function of EXT1. The deletion of EXT1, which is a Golgi-type type II transmembrane glycoprotein, can cause a reduction of HSPGs. HSPGs are an essential component of cartilage and are important regulators of chondrocyte proliferation and differentiation in the growth plate. They are widely distributed on the cell membrane and extracellular matrix of somatic cells. In HME, the disorder of HS synthesis could result in tumor formation. In our study, the deletion of exon 7 was mainly concentrated in patients 1, 2, 3, and 4, and formation of stop codons on exon 7 was mainly concentrated in patients 6 and 8. These mutations were identified in affected individuals and absent in unaffected individuals. Therefore, it is suggested that the change of exon 7 may be related to the pathogenesis of HME. We validated these changes by Sanger sequencing. The deletion mutation and nonsense mutation brought about the loss of 32 amino acid and truncation of EXT1 expression, which may severely disturb the function of EXT1. The results of RT-PCR showed that changes in exon 7 can lead to a 40% reduction in EXT1 gene expression, indicating that the mutation of exon 7 can affect the function of EXT1 to a considerable extent and rapidly promote the process of HME. The mutations exerted a dominant negative effect on the protein encoded by EXT1. As candidate mutations, deletion and nonsense mutations on exon 7 of the EXT1 gene are closely related to HME.
To conclude, this study identified new mutation sites for the pathogenesis of HME, therefore expanding the existing mutation spectrum and providing new targets for the diagnosis and treatment of HME patients. Our findings will support further investigation into the regulation of bone development and tumor formation in HME, as well as the development of new treatments, and will assist genetic counseling of HME.
Acknowledgments
Funding: This work was supported by grants from project of Shanghai Municipal Health Planning commission of China (201740087).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at http://dx.doi.org/10.21037/tp-20-191
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tp-20-191
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tp-20-191). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Ethics Committee of the Department of Orthopedics of Shanghai Children’s Hospital (NO.: 2017023) and informed consent was taken from all the patients.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Schmale GA, Conrad EU 3rd, Raskind WH. The natural history of hereditary multiple exostoses. J Bone Joint Surg Am 1994;76:986-92. [Crossref] [PubMed]
- Wicklund CL, Pauli RM, Johnston D, et al. Natural history study of hereditary multiple exostoses. Am J Med Genet 1995;55:43-6. [Crossref] [PubMed]
- D'Arienzo A, Andreani L, Sacchetti F, et al. Hereditary Multiple Exostoses: Current Insights. Orthop Res Rev 2019;11:199-211. [Crossref] [PubMed]
- Pacifici M. Hereditary Multiple Exostoses: New Insights into Pathogenesis, Clinical Complications, and Potential Treatments. Curr Osteoporos Rep 2017;15:142-52. [Crossref] [PubMed]
- Phan AQ, Pacifici M, Esko JD. Advances in the pathogenesis and possible treatments for multiple hereditary exostoses from the 2016 international MHE conference. Connect Tissue Res 2018;59:85-98. [Crossref] [PubMed]
- Jennes I, Pedrini E, Zuntini M, et al. Multiple osteochondromas: mutation update and description of the multiple osteochondromas mutation database (MOdb). Hum Mutat 2009;30:1620-7. [Crossref] [PubMed]
- Sarrión P, Sangorrin A, Urreizti R, et al. Mutations in the EXT1 and EXT2 genes in Spanish patients with multiple osteochondromas. Sci Rep 2013;3:1346. [Crossref] [PubMed]
- Wuyts W, Bovée JV, Hogendoorn PC. From gene to disease; hereditary multiple exostoses. Ned Tijdschr Geneeskd 2002;146:162-4. [PubMed]
- Wuyts W, Van Hul W, Wauters J, et al. Positional cloning of a gene involved in hereditary multiple exostoses. Hum Mol Genet 1996;5:1547-57. [Crossref] [PubMed]
- Wuyts W, Van Hul W. Molecular basis of multiple exostoses: mutations in the EXT1 and EXT2 genes. Hum Mutat 2000;15:220-7. [Crossref] [PubMed]
- Stancheva-Ivanova MK, Wuyts W, van Hul E, et al. Clinical and molecular studies of EXT1/EXT2 in Bulgaria. J Inherit Metab Dis 2011;34:917-21. [Crossref] [PubMed]
- Pacifici M. The pathogenic roles of heparan sulfate deficiency in hereditary multiple exostoses. Matrix Biol 2018;71-72:28-39. [Crossref] [PubMed]
- Huegel J, Sgariglia F, Enomoto-Iwamoto M, et al. Heparan sulfate in skeletal development, growth, and pathology: the case of hereditary multiple exostoses. Dev Dyn 2013;242:1021-32. [Crossref] [PubMed]
- Oliver GR, Blackburn PR, Ellingson MS, et al. RNA-Seq detects a SAMD12- EXT1 fusion transcript and leads to the discovery of an EXT1 deletion in a child with multiple osteochondromas. Mol Genet Genomic Med 2019;7:e00560. [Crossref] [PubMed]
- Zhuang L, Gerber SD, Kuchen S, et al. Deletion of exon 8 from the EXT1 gene causes multiple osteochondromas (MO) in a family with three affected members. Springerplus 2016;5:71. [Crossref] [PubMed]