
A narrative review of in utero gene therapy: advances, challenges, and future considerations
Introduction
Fetal intervention encompasses an expanding repertoire of treatment strategies including fetal surgery, fetoscopic intervention, stem cell transplant, and in utero gene therapy (IUGT). Fetal surgery and fetoscopic interventions aim to correct anatomic defects and although these techniques could be used to administer IUGT, they are beyond the scope of this review. Additionally, in utero stem cell transplant, specifically hematopoietic stem cell (HSC) transplant, is a well-studied therapeutic modality, but will not be discussed in depth in this review. The focus of this review will be gene therapy strategies utilized in isolation or in combination to better target specific candidate diseases to alleviate or cure disease in utero.
Advances in the fields of Molecular Biology and Maternal Fetal Medicine have led to improved characterization and identification of disease-causing mutations, and their prenatal detection and diagnosis. Clinicians now have several diagnostic techniques that aid in prenatal diagnosis including chorionic villous sampling (CVS), amniocentesis, and detection of cell-free fetal DNA in maternal serum. Following initial screening, CVS or amniocentesis is offered to those pregnancies at increased risk for fetal genetic disorder; however, both tests are considered invasive and carry a risk of loss of pregnancy in approximately 1 in 455 to 900 pregnancies (1). While detection of aneuploidy initially resorted to karyotyping, the development of chromosomal microarray analysis has increased the resolution and number of diseases able to be detected by prenatally (2). In direct comparison, chromosomal microarray analysis had a 6% increase in diagnostic yield over karyotyping in cases of fetal anomaly detected on ultrasound (3). Since 2011, cell-free DNA screening has been commercially available and has decreased utilization of diagnostic procedures. Up to ten percent of circulating DNA in maternal blood originates from the fetal placenta, which cell-free DNA screening uses to amplify and analyze targeted sequences (4). Professional societies have cautioned, however, that cell-free DNA screening remains a screening tool and should not replace confirmatory diagnostic testing (5). With the emergence of genome sequencing, our ability to detect diseases in utero has improved in speed and sensitivity (4,6). However, whole exome sequencing is discouraged in clinical practice without specific indications on prior screening or clinical findings due to false-positive results, low diagnostic yield, high cost, and detection of variations of unknown significance (7).
Now that genetic diseases can be diagnosed in the fetus, IUGT has become a rational therapeutic strategy for monogenetic disease. Under the umbrella of IUGT are technologies that replace or repair the disease-causing mutations to provide the developing fetus with a functional gene product. Examples of candidate diseases include lysosomal storage disorders (LSD), hemoglobinopathies, spinal muscular atrophy (SMA), down syndrome, and cystic fibrosis.
LSD encompass a large group of diseases that affect the metabolism of the lysosome (8). Each individual disease is rare, but collectively, the prevalence of these ranges from 1 in 7,700 to 1 in 5,000 births (8,9). While each LSD is a result of unique mutations in different genes, their resultant deficiencies in enzymatic function lead to the accumulation of toxic metabolites within the lysosome of the cell. Due to the impact of these diseases on the developing brain, there is a compelling rationale to administer therapy before permanent neurologic damage occurs. Replacement of missing enzymes in these disease processes can be limited by immune reactions in postnatal life. Therefore, IUGT represents an ideal opportunity to treat these individuals in utero to develop tolerance to the missing enzymes.
Hemoglobinopathies (sickle cell disease and thalassemias) represent a spectrum of anemia-inducing disorders, attractive for IUGT for their prevalence and morbid manifestation. Sickle cell disease is the most common genetic disease in the United States and affects millions world-wide, resulting in reduced life-expectancy and multi-system organ dysfunction from vaso-occlusive crises (10). Thalassemia causing mutations, while less prevalent in the US than abroad, can be found as variants in up to 5% of the world’s population, clinically impacting up to 70,000 children annually (11). This culminates in fetal anemia, and in the most severe cases, fetal hydrops, endangering the survival of the fetus. Alpha thalassemia major is often fatal in utero without treatment through blood transfusions (12). Beta thalassemia presents as a spectrum of severities classified as minor, intermediate, or major disease. Hundreds of mutations have been identified within the β-globin gene that cause its loss of function. These diseases have deleterious effects on fetal growth and development that can be irreversible. Patients with sickle cell disease, alpha and beta thalassemia who survive to birth may continue to require lifelong treatment with other associated complications such as iron overload and transfusion reactions (13). There is currently a clinical trial to use in utero stem cell transplantation for alpha-thalassemia, as this may reverse the damage caused by the disease in utero and induce tolerance to postnatal boosting stem cell transplantation, however, the fetal bone marrow niche is very competitive and it has proven difficult to attain adequate levels of engraftment (14). IUGT for hemoglobinopathies is attractive because the mutated genes in HSCs could be corrected in situ without the need to compete for the bone marrow cell niche. In particular, the BCL11A gene serves as a single target for gene therapy to potentially benefit patients suffering from hemoglobinopathies affecting the beta subunit of hemoglobin (15). Using this strategy, researchers were able to downregulate expression of BCL11A in order to upregulate expression of fetal hemoglobin within animal models, providing evidence for potential translation (16). It should be noted that alpha thalassemia is commonly caused by a large genetic deletion and is more amenable to cellular therapy than IUGT. However, sickle cell disease and many variants of beta thalassemia, which are commonly caused by single point mutations, would be amenable to IUGT approaches as demonstrated in preclinical animal studies, where in utero gene editing resulted in sustained postnatal correction of anemia.
SMA is a disease characterized by degeneration of the alpha motor neurons resulting in progressive weakness, paralysis, and ultimately death by respiratory failure. One of the main culprits in its disease pathogenesis has been identified as SMN protein encoded by the SMN gene (17). Due to its deleterious effects on patients postnatally, the therapeutic window for this disease occurs early in postnatal life with potential expansion to prenatal life as demonstrated in animal models. Several therapies are now under investigation including delivery of gene therapy via viral vectors within mouse models of SMA. With evidence supporting earlier treatment as beneficial to relieving the SMA phenotype, IUGT is an attractive avenue for further investigation (18-20).
Down syndrome, or Trisomy 21, is the most common trisomy occurring in infants with a prevalence of 6.7 per 10,000 individuals. Children who are born with Down syndrome suffer a spectrum of disability with varying degrees of neurodevelopmental, cardiovascular, and musculoskeletal issues (21). These effects occur early in development and cause lifelong morbidity making Down syndrome an attractive target for IUGT. Mouse models of Down syndrome have allowed researchers to highlight genes such as DYRK1A and their role in neurodevelopment (22). Researchers have targeted DYRK1A in utero using adenoviral vectors to inhibit expression of the gene allowing for partial rescue of neurodevelopmental outcomes in mice. Although these results are promising, clinical trials of Down syndrome treatment have centered on altering the neurotransmitter milieu of the brain through pharmacological means. Studies on efficacy and targeting efficiency are needed to advance preclinical mouse gene therapies to human clinical trials.
Cystic fibrosis is a monogenic disorder that manifests as multisystemic disease from mutations in the cystic fibrosis transmembrane conductance regulator (CFTR) gene. Mutations in CFTR, which encodes a chloride channel, result in the accumulation of thick, dehydrated mucus and occlusion of the conducting airways of the lung, gut lumen, and pancreatic duct. Given the approximate 20% rate of bowel obstruction at birth in the form of meconium ileus and the nearly 80% rate of pancreatic insufficiency at birth, preventing this disease phenotype in utero is a logical and desirable approach (23). Unlike hemoglobinopathies, where HSCs given systemically will pinpoint to the bone marrow niche, cystic fibrosis cannot be treated in this manner as a single corrected stem cell type will not integrate and function in the lung, gut, and pancreas. Systemic IUGT is attractive because it could correct the disease-causing mutation in desired tissues in situ before fetal damage occurs.
Gene therapy can be broadly divided into gene replacement and gene editing strategies. Gene replacement focuses on replacing deficient or missing copies of genes typically using viral vectors. These viral vectors can remain episomal for temporary gene expression or integrate into the host genome providing sustained therapy as shown in Figure 1 (24). The use of viral vectors in postnatal treatment has been limited secondary to immunologic responses. Gene editing focuses on delivering a correct DNA template and utilizing endogenous or exogenous cellular systems to repair incorrect copies of a gene to restore functionality. This approach is viable for short segment deletions, insertions, and single nucleotide polymorphism mutations.
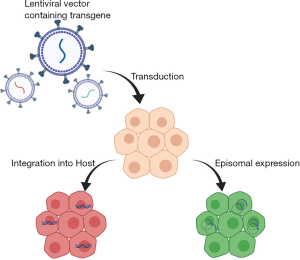
Compared to post-natal gene therapy, the timing of IUGT has several distinct advantages with regard to targeting developmental components of disease processes, the hypo-immune state of the fetus, and access to stem-cell populations. For diseases with a developmental component to the disease phenotype (as described above for LSD, some hemoglobinopathies and cystic fibrosis), gene therapy and correction could take effect prior to the onset of irreversible damage to the developing fetus (25). Another consideration is the hypo-immune state of the fetus. Exploration of adenoviral vectors and adeno-associated viral (AAV) vectors for the postnatal treatment of Duchenne muscular dystrophy have exhibited toxicity attributable to immune responses (26). Initial postnatal human clinical trials of AAV vectors were also noted to elicit responses in both the innate and adaptive immune systems (27,28). Several strategies have been developed to mitigate this immune response. Alterations to the capsid composition, negative selection, and routes of delivery have increased the efficacy of AAV vectors in spite of the host immune system (29). However, shifting the timetable of treatment to in utero may obviate the necessity of these strategies as evidence suggests the nascent fetal immune system develops tolerance to transduced material (25,30-34). Murine models confirm this assertion with an observed increase in engraftment of HSC in immunodeficient mice when compared to immunocompetent mice (25,35,36).
Another distinct advantage to IUGT is the ability to access special populations of cells —specifically stem cells, that are often nascent or inaccessible postnatally (37). To establish sustained expression, IUGT allows for access to stem cell populations within the developing fetus that are generally inaccessible during postnatal life. The progeny of these stem cells divides rapidly in utero and potentiate gene therapy, providing long-term benefits. One aspect that controls specificity of IUGT is vector composition and chemistry. Differences in vectors impart different tropisms for certain cell populations and tissues (25). Due to anatomic features and massive migration and proliferation of fetal stem cell populations, these cells are more susceptible to IUGT and relatively difficult to target after birth (38,39). A related advantage of IUGT is that due to the small size of the fetus treatment is also more efficient because the effective dose per stem cell is dramatically improved compared to postnatally (40).
Delivering gene therapy reagents to the fetus is possible from a technical standpoint in animal models and clinically. Paralleling our ability to detect genetic abnormalities, our ability to provide treatments in utero has advanced. With high-quality ultrasound and technical expertise, delivery of IUGT has become more precise (25,40). Endo et al. described intraamniotic injections as a viable route for delivery of gene therapy, targeting populations of stem cells in both the eye and skin in murine and large animal models (41,42). Direct intramuscular and intraperitoneal injection has also demonstrated efficacy in IUGT deliverance with long-term transgene expression (43,44). Alternatively, intravascular injection into uterine vessels, portal vessels, or yolk sac vessels has demonstrated effective systemic delivery of viral vectors and gene editing reagents in animal models (30,32,40,45). Transitioning to humans, ultrasound-guided umbilical vein access has been the route for fetal transfusion demonstrating feasibility of access for systemic IUGT (46).
We present the following article in accordance with the Narrative Review reporting checklist (available at http://dx.doi.org/10.21037/tp-20-89).
Gene replacement
Gene replacement focuses on delivery of vectors to specific cell populations to help rectify cellular dysfunction caused by absent or deficient gene products. The vectors that deliver the transgenes can be classified as insertional and non-insertional. Lentiviral vectors have utility as insertional gene replacement vectors (Figure 1) (47,48). A notable example of gene replacement using lentiviral vectors is the transduction of HSCs to treat blood dyscrasias in small postnatal clinical trials (49,50). However, in this particular instance, transduction of HSCs occurred ex vivo and HSCs were subsequently re-implanted into the host. While illustrative of gene replacement as a viable strategy, these lentiviral transductions of HSCs occurred in postnatal life subsequent to the timetable for IUGT. There remains a significant body of preclinical work in animal studies demonstrating the propagation of transgene expression after lentiviral transduction of stem cell populations, signifying potential therapeutic applications (38,41,42,44). A disadvantage of insertional viral vectors is the risk of developing malignancies from insertional mutagenesis of tumor suppressor genes (51).
Similarly, adenoviral and AAV vectors have been studied as non-insertional gene replacement solutions. Both have demonstrated efficacy in treating disease models in both small and large animal models. Tran et al. demonstrated stable expression of a reporter gene following in utero transduction of sheep (45). Mattar et al., applying AAV vectors, achieved expression of human recombinant Factor IX in non-human primates that was both sustained and clinically-relevant (30,32). Perhaps the strongest evidence for AAV utilization in gene therapy rests in clinical trials demonstrating efficacy in alleviating hemophilia B and A (52,53). However, these trials were small, conducted in adult men, and no long-term safety conclusions could be definitively drawn from these studies. Adenoviral vectors have been utilized in several clinical trials due to their high transgene expression and tropism for tissues that are prone to adenoviral infection (54). However, limitations exist secondary to hepatotoxicity mediated by cellular immunity (54). Additionally, a notable disadvantage of any viral vector mediated gene replacement strategy is that the gene product is under the control of a non-native promoter sequence that could result in over-expression in an undesired tissue type or under-expression due to silencing.
Gene replacement therapy delivered postnatally overall has low transduction of stem cell populations and low levels of transduction efficiency after systemic delivery. For clotting factor deficiencies such as hemophilia, achieving 1% of the normal level of factor provides therapeutic benefit. However, for diseases like hemoglobinopathies and cystic fibrosis, where 10–25% of normal protein levels are needed, greater levels of expression are needed for clinical benefit.
Gene editing
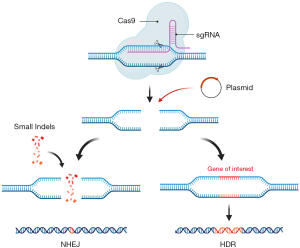
An alternative strategy to gene replacement is gene editing, which can be achieved by endogenous or exogenous nucleases. Gene editing strategies that utilize exogenous nucleases include: zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and CRISPR-Cas9 systems. These modalities either inactivate deleterious genes, correct dysfunctional genes, or alter transcription modifiers to achieve their desired effects. Following cell transduction, nucleases target specific sites to induce double or single stranded breaks in host DNA. Disruption of the host DNA then stimulates the host repair mechanisms to either perform non-homologous end joining, inactivating gene loci through small insertion and deletion (indel) mutations, or homologous recombination with donor DNA.
ZFNs have been used since the 1990s for nuclease-mediated editing in transduced cells. This method couples the FokI endonuclease with a zinc finger domain to target specific sites on host DNA (55). However, each specific gene locus requires a unique construction of a ZFN making each endeavor costly and complex. ZFNs have also demonstrated off-target effects, reflecting a degree of promiscuity in its recognition of nucleotide triplets (55-58). Seeking to improve the efficiency and specificity of genome targeting, both TALENs and CRISPR-Cas systems were developed. Their construction and targeting efficiency catapulted these methods forward for genome editing in both plants and animal models (55). The TALEN system is derived from TALE proteins, discovered in Xanthomonas, which enable binding to specific bases predicted by two critical amino acids within a region of tandem repeats (56). This has made TALENs an attractive modality over ZFNs because it allows researchers to create nucleases specific to certain sequences by altering of the amino acid residues of artificial TALE proteins. Similar to ZFNs, TALENs must dimerize and bind in opposing orientation prior to nuclease activity. In contrast to ZFNs and TALENs, CRISPR-Cas9 functions by providing a non-coding guide RNA in association with Cas protein which has nuclease activity (Figure 2).
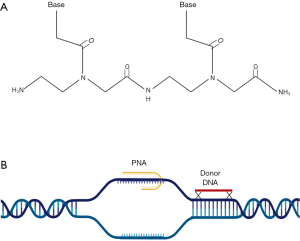
Peptide nucleic acids (PNAs), first described Nielsen et al. in 1991, represent a novel system for gene editing in the absence of exogenous nucleases, with resistance to degradation by endogenous proteases and nucleases (59). PNAs induce gene editing by forming a triple helical structure with a specific DNA target, which signals for endogenous repair mechanisms to mobilize and utilize supplied donor DNA as a template for correction of genetic mutations (Figure 3). Because of the specificity of PNA binding and use of the cell’s own repair mechanisms instead of exogenous nucleases, there are reduced off-target editing, making this method an attractive therapy for systemic clinical therapy (60). Though the backbone of PNA is neutral, its ability to infiltrate cells is limited. Several mechanisms have been proposed including conjugation of PNA to cell-penetrating peptides, but these mechanisms have only demonstrated efficacy in vitro. A solution to this problem was realized when these PNA/DNA agents were loaded into in nanoparticles, which enables systemic delivery and intracellular delivery to their targets (61).
Progress in fetal gene therapy
Significant progress has been achieved over decades of basic and translational research for the use of gene therapy. In vitro and in vivo studies have demonstrated postnatal clinical efficacy and therapies for beta thalassemia, SMA, and an autosomal recessive form of blindness have received formal FDA approval. Rare diseases with no prior treatment options are now experiencing breakthroughs in therapeutic possibilities, with greater than 800 gene therapy programs currently available (62). Therefore, further clinical trials and therapeutic developments are already on their way. Research continues to improve the safety and efficacy of viral vectors, nuclease editing, non-nuclease editing, and combined modalities.
There have been several proof of concept studies demonstrating feasibility of gene replacement therapy in preclinical animal models (some using reporter proteins and others using therapeutic gene products). Rahim et al. described sustained expression of a reporter gene in neural cells following transduction with AAV2/9 in prenatal mice (63). In primates, Binny et al. utilized AAV8 to alleviate congenital Factor VII deficiency in an attempt to prevent early postnatal hemorrhage. The study demonstrated amplification of Factor VII expression follow re-administration of the vector in postnatal life without associated toxicity (64).
Lentiviral vectors have been another popular modality for delivering gene therapy. Small clinical trials in humans have shown potential clinical applications for Wiskott-Aldrich syndrome and metachromatic leukodystrophy; however, long-term safety data remains elusive postnatally (49,50). Another candidate disease being approached via lentiviral vector is Wilson’s disease. Roybal et al. alleviated the complete absence of ATP7B in a knockout murine model with IUGT (65). Studies of IUGT with lentiviral vectors continue to elucidate the timing and route of delivery as it relates to the safety and efficacy of the therapy (41,42).
In recent years there has been progress in testing gene editing strategies in fetal animals. The ability to edit the genome has increased with the advent of CRISPR-Cas9 systems and PNAs. These modalities are ideally suited to target monogenic disorders with small mutations and rely on creating double- or single-stranded breaks to induce endogenous repair mechanisms. Substantial progress has been made with CRISPR-Cas9 in diseases such as hemoglobinopathies and metabolic disorders. The focus of current CRISPR-Cas9 research has been stable delivery of therapeutics to stem cell population where rapid proliferation and gene editing will produce sustained responses postnatally (40). Recently, editing of human HSC ex vivo by Wu et al. allowed for engraftment of corrected human HSC to immunodeficient mice with subsequent alleviation in sickling and an upregulation of fetal hemoglobin (16). Additionally, CRISPR-Cas9 has also been used by Lillegard et al. to perform ex vivo editing in hepatocytes to treat hereditary tyrosinemia in postnatal mice (66). Transitioning to IUGT, Rossidis et al. demonstrated rescue of a lethal phenotype of hereditary tyrosinemia I by adenoviral vector delivery of CRISPR-Cas9 in mice (67).
Two major obstacles for effective IUGT are transducing cells of interest followed by effective and specific gene editing or transgene expression. Packaging PNA/DNA into nanoparticles is another example of combined modality that allows the penetration of specific stem cell populations with active gene editing mechanics. This has been shown to produce thousand-fold increases in gene editing in vivo and in vitro in contrast to naked oligonucleotides (68). In murine models, PNA/DNA packaged into biodegradable polymeric nanoparticles and delivered systemically to the fetus resulted in correction of the beta thalassemia phenotype postnatally and conferred survival advantages over untreated mice. In this study, Ricciardi et al. performed IUGT and were able to access the HSCs to correct dysfunction beta globin genes. Again, this proof of concept study demonstrates the importance of delivering gene therapy in the fetal time period (69).
Future direction and ethical considerations
While IUGT represents a spectrum of exciting therapeutic strategies with curative potential for a range of monogenic disorders, it also comes with a number of ethical considerations. In 2000, the NIH Recombinant DNA Advisory Committee (RAC) issued an executive summary for the research and clinical implementation of IUGT. This position paper established a recommended code of conduct for preclinical and clinical work in gene therapy (70). Further research into optimization of vectors, improvement to accuracy of editing, and reduction of off-target effects will increase the safety and efficacy of existing gene therapies. Although, not applicable in all cases, gene therapy should proceed in larger animal studies to further confirm their safety and efficacy prior to initiation of clinical phase 1 and 2 trials.
One of the most devastating consequences of premature initiation of gene therapies was evidenced by the death of Jesse Gelsinger, who died of overwhelming immune response to an experimental adenoviral vector (71). Arising from this tragic case were several ethical concerns. The safety and efficacy data for gene therapy needed to convey accurate data to patients regarding any potential toxicities. This will ensure that any consent obtained from patients is truly informed. Equally important, researchers must also accurately evaluate the expected harm that may befall a patient. If the risks of harm to a patient are higher than the perceived benefit than regardless of informed consent, the therapy should not proceed. For certain disease processes, arguments can be made that premature fetal death is inevitable without treatment further blurring the line between risks and benefits.
Both the scientific and ethical considerations for IUGT extend beyond the prenatal period into postnatal life. Patient advocacy groups will become increasingly important to bridge the divide between researchers and patients. Researchers and patient advocacy groups should continue to monitor patients well into postnatal life to observe an untoward effect of therapy and promptly make any safety concerns apparent. Part of examining the safety of these therapies, researchers should establish that therapy does not transduce germline stem cells or cause unintended oncogenesis. Several studies have demonstrated that low levels of germline transmission is possible following retroviral gene transfer (72,73). While little is known regarding germline effects, the RAC has concluded that germline integration should be avoided (70). In smaller animal models, the retroviral vectors have demonstrated a series of off-target effects that have resulted in tumorigenesis (51). These effects will be important to monitor in postnatal life following therapy.
Conclusions
Gene therapy provides a range of exciting therapeutic strategies for tackling a number of devastating difficult genetic conditions, many of which have limited or nonexistent postnatal treatments. Application in utero provides distinct advantages over postnatal treatment, but inherently walks the line between disease severity and medical ethics. Research will continue to advance this modality, testing its safety and efficacy on larger animal models and eventually humans. However, it is essential for both patient advocacy groups and researchers to remember that these therapies aim to alleviate disease burden and improve patient quality of life. The balance between scientific progress and patient care must err on the side of patients lest we repeat the tragic lessons of early gene therapy trials. While some diseases, such as hemophilia A or Wilson’s disease, have postnatal treatments available, many monogenetic disorders will rely on advances in IUGT for the development of less costly and more efficient therapeutics.
Acknowledgments
Funding: None.
Footnote
Provenance and Peer Review: This article was commissioned by the Guest Editors (Eric B. Jelin and George B. Mychaliska) for the series “Fetal Surgery” published in Translational Pediatrics. The article was sent for external peer review organized by the Guest Editors and the editorial office.
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at http://dx.doi.org/10.21037/tp-20-89
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tp-20-89). The series “Fetal Surgery” was commissioned by the editorial office without any funding or sponsorship. Dr. ASR has a patent Compositions and Methods for In Utero Delivery pending, a patent Compositions and Methods for Embryonic Gene Editing In Vitro pending, and a patent Compositions and Methods for Enhancing Triplex and Nuclease-Based Gene Editing pending. Dr. DHS has a patent 20200113821 issued. The other authors have no other conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Carlson LM, Vora NL. Prenatal Diagnosis: Screening and Diagnostic Tools. Obstet Gynecol Clin North Am 2017;44:245-56. [Crossref] [PubMed]
- Van den Veyver IB. Recent advances in prenatal genetic screening and testing. F1000Res 2016;5:2591. [Crossref] [PubMed]
- Wapner RJ, Martin CL, Levy B, et al. Chromosomal microarray versus karyotyping for prenatal diagnosis. N Engl J Med 2012;367:2175-84. [Crossref] [PubMed]
- Liao GJ, Lun FM, Zheng YW, et al. Targeted massively parallel sequencing of maternal plasma DNA permits efficient and unbiased detection of fetal alleles. Clin Chem 2011;57:92-101. [Crossref] [PubMed]
- Benn P, Borrell A, Chiu RW, et al. Position statement from the Chromosome Abnormality Screening Committee on behalf of the Board of the International Society for Prenatal Diagnosis. Prenat Diagn 2015;35:725-34. [Crossref] [PubMed]
- Kitzman JO, Snyder MW, Ventura M, et al. Noninvasive whole-genome sequencing of a human fetus. Sci Transl Med 2012;4:137ra76 [Crossref] [PubMed]
- Jelin AC, Vora N. Whole Exome Sequencing: Applications in Prenatal Genetics. Obstet Gynecol Clin North Am 2018;45:69-81. [Crossref] [PubMed]
- Meikle PJ, Hopwood JJ, Clague AE, et al. Prevalence of lysosomal storage disorders. JAMA 1999;281:249-54. [Crossref] [PubMed]
- Fuller M, Meikle PJ, Hopwood JJ. Epidemiology of lysosomal storage diseases: an overview. In: Mehta A, Beck M, Sunder-Plassmann G. editors. Fabry Disease: Perspectives from 5 Years of FOS. Oxford, 2006.
- Hoppe C, Neumayr L. Sickle Cell Disease: Monitoring, Current Treatment, and Therapeutics Under Development. Hematol Oncol Clin North Am 2019;33:355-71. [Crossref] [PubMed]
- Martin A, Thompson AA. Thalassemias. Pediatr Clin North Am 2013;60:1383-91. [Crossref] [PubMed]
- Han XD, Lin C, Chang J, et al. Fetal gene therapy of alpha-thalassemia in a mouse model. Proc Natl Acad Sci U S A 2007;104:9007-11. [Crossref] [PubMed]
- Raja JV, Rachchh MA, Gokani RH. Recent advances in gene therapy for thalassemia. J Pharm Bioallied Sci 2012;4:194-201. [Crossref] [PubMed]
- Mackenzie TC, Shaaban AF, Radu A, et al. Engraftment of bone marrow and fetal liver cells after in utero transplantation in MDX mice. J Pediatr Surg 2002;37:1058-64. [Crossref] [PubMed]
- Humbert O, Peterson CW, Norgaard ZK, et al. A Nonhuman Primate Transplantation Model to Evaluate Hematopoietic Stem Cell Gene Editing Strategies for beta-Hemoglobinopathies. Mol Ther Methods Clin Dev 2017;8:75-86. [Crossref] [PubMed]
- Wu Y, Zeng J, Roscoe BP, et al. Highly efficient therapeutic gene editing of human hematopoietic stem cells. Nat Med 2019;25:776-83. [Crossref] [PubMed]
- Zanetta C, Riboldi G, Nizzardo M, et al. Molecular, genetic and stem cell-mediated therapeutic strategies for spinal muscular atrophy (SMA). J Cell Mol Med 2014;18:187-96. [Crossref] [PubMed]
- Lutz CM, Kariya S, Patruni S, et al. Postsymptomatic restoration of SMN rescues the disease phenotype in a mouse model of severe spinal muscular atrophy. J Clin Invest 2011;121:3029-41. [Crossref] [PubMed]
- Farrar MA, Park SB, Vucic S, et al. Emerging therapies and challenges in spinal muscular atrophy. Ann Neurol 2017;81:355-68. [Crossref] [PubMed]
- Phan HC, Taylor JL, Hannon H, et al. Newborn screening for spinal muscular atrophy: Anticipating an imminent need. Semin Perinatol 2015;39:217-29. [Crossref] [PubMed]
- Antonarakis SE, Skotko BG, Rafii MS, et al. Down syndrome. Nat Rev Dis Primers 2020;6:9. [Crossref] [PubMed]
- Guedj F, Bianchi DW, Delabar JM. Prenatal treatment of Down syndrome: a reality? Curr Opin Obstet Gynecol 2014;26:92-103. [Crossref] [PubMed]
- Davies JC, Alton EWFW, Bush A. Cystic fibrosis. BMJ 2007;335:1255-9. [Crossref] [PubMed]
- Waddington SN, Kramer MG, Hernandez-Alcoceba R, et al. In utero gene therapy: current challenges and perspectives. Mol Ther 2005;11:661-76. [Crossref] [PubMed]
- Almeida-Porada G, Atala A, Porada CD. In utero stem cell transplantation and gene therapy: rationale, history, and recent advances toward clinical application. Mol Ther Methods Clin Dev 2016;5:16020. [Crossref] [PubMed]
- Duan D. Systemic AAV Micro-dystrophin Gene Therapy for Duchenne Muscular Dystrophy. Mol Ther 2018;26:2337-56. [Crossref] [PubMed]
- Basner-Tschakarjan E, Bijjiga E, Martino AT. Pre-Clinical Assessment of Immune Responses to Adeno-Associated Virus (AAV) Vectors. Front Immunol 2014;5:28. [Crossref] [PubMed]
- Bessis N. GarciaCozar FJ, Boissier MC. Immune responses to gene therapy vectors: influence on vector function and effector mechanisms. Gene Ther 2004;11:S10-7. [Crossref] [PubMed]
- Barnes C, Scheideler O, Schaffer D. Engineering the AAV capsid to evade immune responses. Curr Opin Biotechnol 2019;60:99-103. [Crossref] [PubMed]
- Mattar CNZ, Gil-Farina I, Rosales C, et al. In Utero Transfer of Adeno-Associated Viral Vectors Produces Long-Term Factor IX Levels in a Cynomolgus Macaque Model. Mol Ther 2017;25:1843-53. [Crossref] [PubMed]
- Billingham RE, Brent L, Medawar PB. Actively acquired tolerance of foreign cells. Nature 1953;172:603-6. [Crossref] [PubMed]
- Mattar CN, Nathwani AC, Waddington SN, et al. Stable human FIX expression after 0.9G intrauterine gene transfer of self-complementary adeno-associated viral vector 5 and 8 in macaques. Mol Ther 2011;19:1950-60. [Crossref] [PubMed]
- Mold JE, Michaelsson J, Burt TD, et al. Maternal alloantigens promote the development of tolerogenic fetal regulatory T cells in utero. Science 2008;322:1562-5. [Crossref] [PubMed]
- Mold JE, Venkatasubrahmanyam S, Burt TD, et al. Fetal and adult hematopoietic stem cells give rise to distinct T cell lineages in humans. Science 2010;330:1695-9. [Crossref] [PubMed]
- Nijagal A, Wegorzewska M, Jarvis E, et al. Maternal T cells limit engraftment after in utero hematopoietic cell transplantation in mice. J Clin Invest 2011;121:582-92. [Crossref] [PubMed]
- Nijagal A, Derderian C, Le T, et al. Direct and indirect antigen presentation lead to deletion of donor-specific T cells after in utero hematopoietic cell transplantation in mice. Blood 2013;121:4595-602. [Crossref] [PubMed]
- Waddington SN, Buckley SM, Nivsarkar M, et al. In utero gene transfer of human factor IX to fetal mice can induce postnatal tolerance of the exogenous clotting factor. Blood 2003;101:1359-66. [Crossref] [PubMed]
- Endo M, Henriques-Coelho T, Zoltick PW, et al. The developmental stage determines the distribution and duration of gene expression after early intra-amniotic gene transfer using lentiviral vectors. Gene Ther 2010;17:61-71. [Crossref] [PubMed]
- Karda R, Buckley SM, Mattar CN, et al. Perinatal systemic gene delivery using adeno-associated viral vectors. Front Mol Neurosci 2014;7:89. [Crossref] [PubMed]
- Witt R, MacKenzie TC, Peranteau WH. Fetal stem cell and gene therapy. Semin Fetal Neonatal Med 2017;22:410-4. [Crossref] [PubMed]
- Endo M, Zoltick PW, Chung DC, et al. Gene transfer to ocular stem cells by early gestational intraamniotic injection of lentiviral vector. Mol Ther 2007;15:579-87. [Crossref] [PubMed]
- Endo M, Zoltick PW, Peranteau WH, et al. Efficient in vivo targeting of epidermal stem cells by early gestational intraamniotic injection of lentiviral vector driven by the keratin 5 promoter. Mol Ther 2008;16:131-7. [Crossref] [PubMed]
- Bouchard S, MacKenzie TC, Radu AP, et al. Long-term transgene expression in cardiac and skeletal muscle following fetal administration of adenoviral or adeno-associated viral vectors in mice. J Gene Med 2003;5:941-50. [Crossref] [PubMed]
- MacKenzie TC, Kobinger GP, Louboutin JP, et al. Transduction of satellite cells after prenatal intramuscular administration of lentiviral vectors. J Gene Med 2005;7:50-8. [Crossref] [PubMed]
- Tran ND, Porada CD, Almeida-Porada G, et al. Induction of stable prenatal tolerance to beta-galactosidase by in utero gene transfer into preimmune sheep fetuses. Blood 2001;97:3417-23. [Crossref] [PubMed]
- Lindenburg IT, van Kamp IL, Oepkes D. Intrauterine blood transfusion: current indications and associated risks. Fetal Diagn Ther 2014;36:263-71. [Crossref] [PubMed]
- Kumar M, Bradow BP, Zimmerberg J. Large-scale production of pseudotyped lentiviral vectors using baculovirus GP64. Hum Gene Ther 2003;14:67-77. [Crossref] [PubMed]
- Schauber CA, Tuerk MJ, Pacheco CD, et al. Lentiviral vectors pseudotyped with baculovirus gp64 efficiently transduce mouse cells in vivo and show tropism restriction against hematopoietic cell types in vitro. Gene Ther 2004;11:266-75. [Crossref] [PubMed]
- Aiuti A, Biasco L, Scaramuzza S, et al. Lentiviral hematopoietic stem cell gene therapy in patients with Wiskott-Aldrich syndrome. Science 2013;341:1233151 [Crossref] [PubMed]
- Biffi A, Montini E, Lorioli L, et al. Lentiviral hematopoietic stem cell gene therapy benefits metachromatic leukodystrophy. Science 2013;341:1233158 [Crossref] [PubMed]
- Themis M, Waddington SN, Schmidt M, et al. Oncogenesis following delivery of a nonprimate lentiviral gene therapy vector to fetal and neonatal mice. Mol Ther 2005;12:763-71. [Crossref] [PubMed]
- Rangarajan S, Walsh L, Lester W, et al. AAV5-Factor VIII Gene Transfer in Severe Hemophilia A. N Engl J Med 2017;377:2519-30. [Crossref] [PubMed]
- George LA, Sullivan SK, Giermasz A, et al. Hemophilia B Gene Therapy with a High-Specific-Activity Factor IX Variant. N Engl J Med 2017;377:2215-27. [Crossref] [PubMed]
- Douglas JT. Adenoviral vectors for gene therapy. Mol Biotechnol 2007;36:71-80. [Crossref] [PubMed]
- Nemudryi AA, Valetdinova KR, Medvedev SP, et al. TALEN and CRISPR/Cas Genome Editing Systems: Tools of Discovery. Acta Naturae 2014;6:19-40. [Crossref] [PubMed]
- Miller JC, Tan S, Qiao G, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol 2011;29:143-8. [Crossref] [PubMed]
- Carroll D. Progress and prospects: zinc-finger nucleases as gene therapy agents. Gene Ther 2008;15:1463-8. [Crossref] [PubMed]
- Chou ST, Leng Q, Mixson AJ. Zinc Finger Nucleases: Tailor-made for Gene Therapy. Drugs Future 2012;37:183-96. [Crossref] [PubMed]
- Nielsen PE, Egholm M, Berg RH, et al. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 1991;254:1497-500. [Crossref] [PubMed]
- Ricciardi AS, Quijano E, Putman R, et al. Peptide Nucleic Acids as a Tool for Site-Specific Gene Editing. Molecules 2018;23:632. [Crossref] [PubMed]
- Quijano E, Bahal R, Ricciardi A, et al. Therapeutic Peptide Nucleic Acids: Principles, Limitations, and Opportunities. Yale J Biol Med 2017;90:583-98. [PubMed]
- High KA, Roncarolo MG. Gene Therapy. N Engl J Med 2019;381:455-64. [Crossref] [PubMed]
- Rahim AA, Wong AM, Ahmadi S, et al. In utero administration of Ad5 and AAV pseudotypes to the fetal brain leads to efficient, widespread and long-term gene expression. Gene Ther 2012;19:936-46. [Crossref] [PubMed]
- Binny C, McIntosh J, Della Peruta M, et al. AAV-mediated gene transfer in the perinatal period results in expression of FVII at levels that protect against fatal spontaneous hemorrhage. Blood 2012;119:957-66. [Crossref] [PubMed]
- Roybal JL, Endo M, Radu A, et al. Early gestational gene transfer with targeted ATP7B expression in the liver improves phenotype in a murine model of Wilson's disease. Gene Ther 2012;19:1085-94. [Crossref] [PubMed]
- VanLith CJ, Guthman RM, Nicolas CT, et al. Ex Vivo Hepatocyte Reprograming Promotes Homology-Directed DNA Repair to Correct Metabolic Disease in Mice After Transplantation. Hepatol Commun 2019;3:558-73. [Crossref] [PubMed]
- Rossidis AC, Stratigis JD, Chadwick AC, et al. In utero CRISPR-mediated therapeutic editing of metabolic genes. Nat Med 2018;24:1513-8. [Crossref] [PubMed]
- McNeer NA, Schleifman EB, Cuthbert A, et al. Systemic delivery of triplex-forming PNA and donor DNA by nanoparticles mediates site-specific genome editing of human hematopoietic cells in vivo. Gene Ther 2013;20:658-69. [Crossref] [PubMed]
- Ricciardi AS, Bahal R, Farrelly JS, et al. In utero nanoparticle delivery for site-specific genome editing. Nat Commun 2018;9:2481. [Crossref] [PubMed]
- U.S. National Institutes of Health. Prenatal gene tranfer: scientific, medical, and ethical issues: a report of the Recombinant DNA Advisory Committee. Hum Gene Ther 2000;11:1211-29. [Crossref] [PubMed]
- Savulescu J. Harm, ethics committees and the gene therapy death. J Med Ethics 2001;27:148-50. [Crossref] [PubMed]
- Porada CD, Park PJ, Tellez J, et al. Male germ-line cells are at risk following direct-injection retroviral-mediated gene transfer in utero. Mol Ther 2005;12:754-62. [Crossref] [PubMed]
- Park PJ, Colletti E, Ozturk F, et al. Factors determining the risk of inadvertent retroviral transduction of male germ cells after in utero gene transfer in sheep. Hum Gene Ther 2009;20:201-15. [Crossref] [PubMed]


