
Clinical exome sequencing revealed that FLNC variants contribute to the early diagnosis of cardiomyopathies in infant patients
Introduction
FLNC is the coding gene of filamin C, and is an actin-binding protein, mainly concentrated in skeletal and cardiac muscle (1). FLNC is distributed in the Z disk and the sarcomere in the formation of muscle, suggesting that the gene is involved in sarcomere formation (2). The FLNC gene is located in chr7q32-q35, comprises 47 exons, and belongs to the filamin family. This family, including Filamin A, Filamin B and Filamin C, are large molecule proteins mainly distributed in the cytoplasm, where they maintain the three-dimensional structure of cortical actin cross-linking protein and play an important role in the adhesion between cells and cell and interstitial and intracellular signaling pathways (3). FLNA, FLNB and FLNC genes are highly conserved and widely expressed in the development process. FLNC mutation was first found in patients with filamin associated myofibrillar myopathy (4).
In 2014, Valdes-Mas et al. found by exome sequencing that A1539T in FLNC in a family segregates with hereditary cardiomyopathy. Since then, approximately 90 mutations in FLNC have been found in cardiomyopathies, including restrictive cardiomyopathy (RCM) (5), hypertrophic cardiomyopathy (HCM) (6), and dilated cardiomyopathy (DCM) (7). These mutations can be divided into the following types: missense, splicing, small deletions, small insertions, small indels, gross deletions, and gross insertions. To date, the genetic roles of FLNC mutations have mainly been reported in adults. There are limited studies investigating the relationship between FLNC variants and cardiac phenotypes in pediatric patients.
In the present study, we summarized the variants of FLNC in pediatric patients who have the phenotype of cardiomyopathy from a large cohort of clinical exome sequencing (CES) data to identify FLNC variants in patients with cardiomyopathy. We sought to provide new variants and more evidence for FLNC variants in pediatric patients.
Methods
Ethical compliance
The criteria for genetic testing were approved by the ethics committee of Children’s Hospital of Fudan University (2016-235). Pretest counseling was performed by physicians, and appropriate informed consent was signed by the patient’s parents in clinics. The data we analyzed were from pediatric patients with cardiomyopathy who underwent CES from May 2016 to May 2019 in the Center for Molecular Medicine, Children’s Hospital of Fudan University and the Maternal & Child Health Hospital of Guangxi Zhuang Autonomous Region. The clinical manifestations and phenotype data were collected from medical record, and family members’ blood was collected for parental Sanger confirmation.
Clinical exome sequencing
Genomic DNA was extracted from peripheral blood using the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. High-throughput sequencing was performed using standard protocols in the Clinical Laboratory Improvement Amendments compliant sequencing laboratory in Wuxi NEXTCODE (288 Fute Zhong Road, Waigaoqiao Free Trade Zone Shanghai 200131, China CLIA ID 99D2064856). The Agilent ClearSeq Inherited Disease Kit Illumina Cluster and SBS kit were used for clinical exome sequencing. Sequencing was performed using next-generation sequencing on an Illumina HiSeq 2000/2500 platform. It can cover a minimum of 98% of the genome with 20X coverage or higher. Clean reads were aligned to the reference human genome (UCSC hg19), and variant calling was performed using the Genome Analysis Tool Kit, (GATK) v.2.3-9 by applying base quality score recalibration, indel realignment and performing SNP and INDEL discovery and genotyping. The three major databases for known pathogenic and likely pathogenic mutations were used including ClinVar, Online Mendelian Inheritance in Man (OMIM), and the Human Gene Mutation Database (HGMD, professional version). Numerous missense functional predictors, noncoding regulatory region annotations, and others were included. To filter out variants with high frequency, the allelic frequencies were annotated from the public datasets from the 1000 Genomes, the Exome Sequencing Project (EVS6500) and Exome Aggregation Consortium (ExAC), Kyoto, and Dutch allelic frequency databases. Data interpretation followed our previous work (8). All aligned reads, variants, and annotations were stored according to the genomic position in the WuXi NextCODE Genomically Ordered Relational Database (GORdb) to facilitate more rapid access by the Clinical Sequence Analyzer (CSA) user interface, Sequence Miner, and the WuXi NextCODE Genome Browser.
Sanger sequencing
Variants of patient and parents were validated by Sanger sequencing using an automated sequencer (3500XL Genetic Analyzer, Applied Biosystems, USA). Primers were designed for polymerase chain reaction amplification using Primer Premier 5.0 software. Sequence analysis was performed using MutationSurveyor (SoftGenetics®, State College, PA, USA).
Protein structure analysis
The structures of FLNC in wild type and mutants were analyzed by UniProt (http://www.uniprot.org/), Mutalyzer (https://www.mutalyzer.nl/), and SWISS-MODEL (https://www.swissmodel.expasy.org). The FASTA sequences of FLNC from UniProt were downloaded. We selected 3 templates, including 6ew1.1 (9), 2j3s.1 (10), and 4m9p.1 (11), as the protein structure template. We used a computer program MODELLER (12) for automated modeling. Protein structures were visualized by PyMOL 2.3.
Results
Patient cohort
The present study enrolled 122 patients with cardiomyopathy for whom CES was conducted. Five patients were identified with FLNC variants, accounting for 4.1% (5/122) of all the enrolled patients.
Clinical characteristics
Five cases (3 males and 2 females) with cardiomyopathy characterized by similar phenotypes of severe malformations in the cardiac phenotypes were summarized. The median age was 3 months (range: 19 days to 30 months). All patients had no family histories. The clinical characteristics of each patient are shown in Table 1. Patient 1 and patient 3 were diagnosed with RCM. Patient 2 and patient 5 were diagnosed with DCM. Patient 4 was diagnosed with arrhythmia. All of them had an abnormal echocardiography (ECHO) result. All patients survived up to the last follow-up.
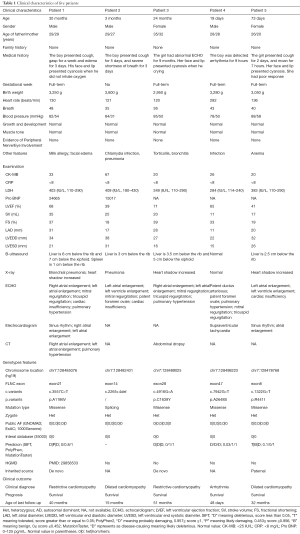
Full table
Patient 1 was a 2-year-6-month-old boy who was delivered after a full-term pregnancy and had no history of suffocation and rescue at birth. The patient presented cough and gasping for 1 week. ECHO showed that he had right atrial enlargement, left atrial enlargement, mitral regurgitation, tricuspid regurgitation, cardiac insufficiency, and pulmonary hypertension. Serum biochemistry tests showed high levels of pro-brain natriuretic peptide (Pro-BNP; 24,655 pg/mL; normal range <125 pg/mL) and LDH (403 mg/L; normal range: 110–290 IU/L).
Patient 2 was a 3-month-3-day-old boy. His delivery is normal and had no history of suffocation and rescue at birth. He presented to our hospital for severe shortness of breath at 3 months. ECHO showed that he had left atrial enlargement, left ventricle enlargement, mitral regurgitation, patent foramen ovale (PFO), and cardiac insufficiency. Serum biochemistry tests showed high levels of Pro-BNP (15,017 pg/mL).
Patient 3 was a 2-year-old girl. She was born by cesarean section. At 15 months of age, the ECHO showed that she had pulmonary hypertension. The girl breaths faster after slight activities and presents with perioral cyanosis when crying. She was transferred to our hospital for precision treatment. The results of ECHO in our hospital showed that she had atrial enlargement, mitral regurgitation, tricuspid regurgitation, and pulmonary hypertension.
Patient 4 was a 19-day-old boy who was born via cesarean section. He had no history of suffocation and rescue at birth. The electrocardiogram showed that the boy had arrhythmia in routine medical examinations. His heart rate was 282 beats/min. ECHO results showed that he had patent ductus arteriosus (PDA), PFO, pulmonary hypertension, mitral regurgitation, and tricuspid regurgitation. Electrocardiogram (EKG) showed that he had supraventricular tachycardia.
Patient 5 was a 2-month-7-day-old girl who was delivered after a full-term pregnancy and had no history of suffocation and rescue at birth. The girl presented cough for 2 days and moaned for 7 hours. ECHO showed that she had left atrial enlargement, left ventricle enlargement, and cardiac insufficiency.
Genetic testing results
The CES data were analyzed following the clinical genetic analysis pipeline developed by our teams (8,13). Five patients were diagnosed with FLNC mutations. One known pathogenic variant (c.3557C>T, A1186V) and four novel variants (c.2265+4del, p.R441I, p.C1639Y, and p.A2648S) were identified in this study (Table 1, Figure 1). A1186V found in patient 1 was reported in the previous study (14), and in three patients with RCM, and their ages were 1.4 years, 3 years, and 15 years old. A1186V was recorded as a pathogenic variant in ClinVar and the Human Gene Mutation Database (HGMD). One splicing variant (c.2265+4del) was identified in patient 2 and three novel missense variants were identified in three individuals (c.4916G>A, p.C1639Y in patient 3; c.7942G>T, p.A2648S in patient 4; c.1322G>T, p.R441I in patient 5).
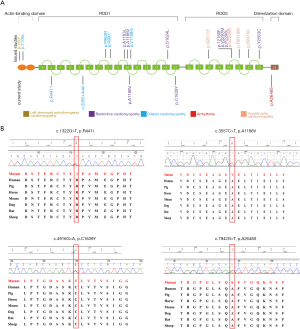
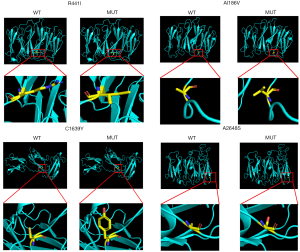
The four novel variants were not found in known public databases (GnomAD, ExAC, and 1000Genome) and internal database (more than 30,000 CES samples). Except for A2648S in the dimerization domain, others are located in the Ig domain. All missense variants are highly conserved in different species. The prediction results of SIFT, PolyPhen, and MutationTaster were harmful. Detailed information is shown in Table 1, and the predicted structures are shown in Figure 2.
Discussion
In 2005, a disease-related mutation within the FLNC gene was first described in myofibrillar myopathy by Vorgerd et al. (4), and more reports considered FLNC to be related to myofibrillar myopathies (15-17). In 2014, Valdes-Mas et al. reported FLNC in the HCM family (p.A1539T) (6). Since then, ten more studies have described FLNC mutations associated with cardiomyopathy in different races, including Caucasian and Chinese populations (5,18,19). Recently, the Mayo Clinic reported that a novel truncate variant in FLNC may serve as a proarrhythmic genetic substrate for arrhythmogenic bileaflet mitral valve prolapse syndrome in a 51-year-old man and his relatives with similar symptoms (20). The author regarded that this genetic substrate may underlie, at least in part, some cases of arrhythmogenic mitral valve prolapse syndrome. In addition, FLNC mutations are associated with high-risk dilated and arrhythmogenic cardiomyopathies, and cause sudden cardiac death (7,21). However, only 24 patients with 16 types of mutations in the FLNC-associated cardiac phenotype have been reported, which disappears when leaving most of them become adults (5,19,21,22).
In this study, we included 5 pediatric cases displaying a FLNC-associated cardiac phenotype. The spectrum of FLNC-associated phenotypes includes RCM, arrhythmia, and DCM. The missense variant (R441I) in patient 5 with DCM is paternally inherited. When followed up by telephone, the patient’s father (26 years old) did not currently complain of heart problems. A previous study showed that FLNC variants such as E637G, R1150Q, and T2099N can be found in patients with HCM or in healthy individuals (18), indicating incomplete penetrance. Valdes-Mas et al. (6) reported that, with 14 of 16 mutation carriers over 40 years of age exhibiting symptoms of HCM, the penetrance is more than 87%. The father of patient 5 carrying R441I maybe be due to incompletely penetrance or later onset cardiomyopathy.
A search of the PubMed database revealed a total of 17 publications reporting FLNC variants related to 294 patients with cardiac phenotypes. Of these, 7 family studies reported 16 mutations in 24 pediatric patients with DCM, RCM, HCM, or left-dominant arrhythmogenic cardiomyopathy (5,14,19,21-24). Of these, 10 were male and 14 were female. The diagnosed age ranged from 3 months to 17 years old (Table 2). Among all the reported FLNC induced cardiomyopathies, there are only four patients diagnosed with RCM accompanied by a CHD phenotype (5,14). Thus, CHD may be an early onset phenotype accompanied by cardiomyopathy. In this study, a rare novel FLNC variant (A2648S) was found in patient 4. Notably, arrhythmia, a symptom of cardiomyopathy (25), was identified by EKG in this patient. Moreover, ECHO observed that he had PDA and PFO, but not cardiomyopathy due to young age (19 days). We think this patient may progress to cardiomyopathy with age. Therefore, our early diagnosis can provide an opportunity for early comprehensive intervention that delays other cardiac phenotypes disease progression.
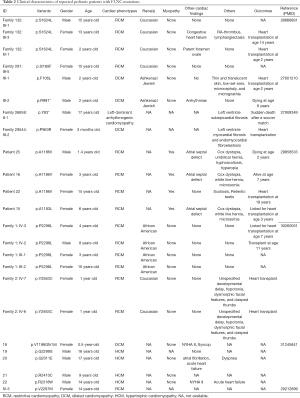
Full table
Recently, several studies have focused on the relationship between the FLNC genotype and imaging phenotype. Ader et al. (24) found no particular association between the rhythm disturbance and the position of FLNC variants in 28 patients with cardiomyopathies. However, Sveinbjornsson et al. (7) indicated that the FLNC variant in DCM patients similarly affects many ECG traits, with the most notable association with the R-wave amplitude. In addition, the HCM patients with and without FLNC mutations had similar left ventricular ejection fraction features (18). Interestingly, a similar result was reported by Augusto et al. (26). Moreover, they conducted a comprehensive genotype-imaging phenotype study and found that DSP/FLNC genotypes cause more regionality in left ventricular impairment in DCM patients. Thus, FLNC variations may account for different imaging phenotypes compared with other gene mutations. In the current study, we did not conduct genotype-imaging phenotype analysis because only 5 patients were included. We will explore the relationship between genotype and imaging phenotype in future studies.
More interestingly, the variant (p.A2648S) found in this patient is located within the dimerization domain which results in the inability to dimerize properly and renders the protein more susceptible to proteolysis (4,27). This phenomenon may be related to myocardial development and may cause cardiomyopathy. However, further experiments with more patient samples are needed to confirm this hypothesis. The remaining four variants are located within ROD1, which can lead to filamin aggregates (15). In addition, based on protein structure analysis, we found that one variant made the amino acid more hydrophobic (A1186V), one variant changed the hydrophilic amino acid into a hydrophobic amino acid (R441I), and two variants (C1639Y and A2648S) changed the hydrophobic amino acids into hydrophilic amino acids (Figure 2). Combined with the prediction of the variants by in silico tools (MutationTaster, PolyPhen, and SIFT), the prediction results may illustrate pathogenesis. Further functional studies are needed in the future.
Conclusions
In summary, the FLNC variant was identified as an early diagnosis genetic alteration associated with cardiomyopathy occurrence in infant patients. This research expanded the mutation spectrum of the FLNC gene.
Acknowledgments
We are very grateful to the patient families and thanks for their trust for our lab. We thank clinician for taking care of the patient, and our genetic laboratory teams who contributed to this study.
Funding: This work was supported by the National Natural Science Foundation of China [81471483], and the Shanghai Key Laboratory of Birth Defects [13DZ2260600].
Footnote
Conflict of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The criteria of genetic testing were got approved of the ethics committee of Children’s Hospital of Fudan University (2016-235). Pre-test counseling was performed by physicians, appropriate informed consent was signed by the patient’s parents in clinics.
References
- Thompson TG, Chan YM, Hack AA, et al. Filamin 2 (FLN2): A muscle-specific sarcoglycan interacting protein. J Cell Biol 2000;148:115-26. [Crossref] [PubMed]
- van der Ven PF, Obermann WM, Lemke B, et al. Characterization of muscle filamin isoforms suggests a possible role of gamma-filamin/ABP-L in sarcomeric Z-disc formation. Cell Motil Cytoskeleton 2000;45:149-62. [Crossref] [PubMed]
- Weihing RR. The filamins: properties and functions. Can J Biochem Cell Biol 1985;63:397-413. [Crossref] [PubMed]
- Vorgerd M, van der Ven PF, Bruchertseifer V, et al. A mutation in the dimerization domain of filamin c causes a novel type of autosomal dominant myofibrillar myopathy. Am J Hum Genet 2005;77:297-304. [Crossref] [PubMed]
- Brodehl A, Ferrier RA, Hamilton SJ, et al. Mutations in FLNC are Associated with Familial Restrictive Cardiomyopathy. Hum Mutat 2016;37:269-79. [Crossref] [PubMed]
- Valdés-Mas R, Gutierrez-Fernandez A, Gomez J, et al. Mutations in filamin C cause a new form of familial hypertrophic cardiomyopathy. Nat Commun 2014;5:5326. [Crossref] [PubMed]
- Sveinbjornsson G, Olafsdottir EF, Thorolfsdottir RB, et al. Variants in NKX2-5 and FLNC Cause Dilated Cardiomyopathy and Sudden Cardiac Death. Circ Genom Precis Med 2018;11:e002151. [Crossref] [PubMed]
- Yang L, Kong Y, Dong X, et al. Clinical and genetic spectrum of a large cohort of children with epilepsy in China. Genet Med 2019;21:564-71. [Crossref] [PubMed]
- Haataja TJK, Bernardi RC, Lecointe S, et al. Non-syndromic Mitral Valve Dysplasia Mutation Changes the Force Resilience and Interaction of Human Filamin A. Structure 2019;27:102-12.e4. [Crossref] [PubMed]
- Lad Y, Kiema T, Jiang P, et al. Structure of three tandem filamin domains reveals auto-inhibition of ligand binding. Embo j 2007;26:3993-4004. [Crossref] [PubMed]
- Sethi R, Seppala J, Tossavainen H, et al. A novel structural unit in the N-terminal region of filamins. J Biol Chem 2014;289:8588-98. [Crossref] [PubMed]
- Martí-Renom MA, Stuart AC, Fiser A, et al. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct 2000;29:291-325. [Crossref] [PubMed]
- Li Z, Zhang F, Wang Y, et al. PhenoPro: a novel toolkit for assisting in the diagnosis of Mendelian disease. Bioinformatics 2019;35:3559-66. [Crossref] [PubMed]
- Kiselev A, Vaz R, Knyazeva A. De novo mutations in FLNC leading to early-onset restrictive cardiomyopathy and congenital myopathy. Hum Mutat 2018;39:1161-72. [Crossref] [PubMed]
- Kley RA, Serdaroglu-Oflazer P, Leber Y, et al. Pathophysiology of protein aggregation and extended phenotyping in filaminopathy. Brain 2012;135:2642-60. [Crossref] [PubMed]
- Miao J, Su FF, Liu XM, et al. A case report: a heterozygous deletion (2791_2805 del) in exon 18 of the filamin C gene causing filamin C-related myofibrillar myopathies in a Chinese family. BMC Neurol 2018;18:79. [Crossref] [PubMed]
- Gemelli C, Prada V, Fiorillo C, et al. A novel mutation in the N-terminal acting-binding domain of Filamin C protein causing a distal myofibrillar myopathy. J Neurol Sci 2019;398:75-8. [Crossref] [PubMed]
- Cui H, Wang J, Zhang C, et al. Mutation profile of FLNC gene and its prognostic relevance in patients with hypertrophic cardiomyopathy. Mol Genet Genomic Med 2018;6:1104-13. [Crossref] [PubMed]
- Schubert J, Tariq M, Geddes G, et al. Novel pathogenic variants in filamin C identified in pediatric restrictive cardiomyopathy. Hum Mutat 2018;39:2083-96. [Crossref] [PubMed]
- Bains S, Tester DJ, Asirvatham SJ, et al. A Novel Truncating Variant in FLNC-Encoded Filamin C May Serve as a Proarrhythmic Genetic Substrate for Arrhythmogenic Bileaflet Mitral Valve Prolapse Syndrome. Mayo Clin Proc 2019;94:906-13. [Crossref] [PubMed]
- Ortiz-Genga MF, Cuenca S, Dal Ferro M, et al. Truncating FLNC Mutations Are Associated With High-Risk Dilated and Arrhythmogenic Cardiomyopathies. J Am Coll Cardiol 2016;68:2440-51. [Crossref] [PubMed]
- Reinstein E, Gutierrez-Fernandez A, Tzur S, et al. Congenital dilated cardiomyopathy caused by biallelic mutations in Filamin C. Eur J Hum Genet 2016;24:1792-6. [Crossref] [PubMed]
- Tucker NR, McLellan MA, Hu D, et al. Novel Mutation in FLNC (Filamin C) Causes Familial Restrictive Cardiomyopathy. Circ Cardiovasc Genet 2017;10. [Crossref] [PubMed]
- Ader F, De Groote P, Reant P, et al. FLNC pathogenic variants in patients with cardiomyopathies: Prevalence and genotype-phenotype correlations. Clin Genet 2019;96:317-29. [Crossref] [PubMed]
- Lee TM, Hsu DT, Kantor P, et al. Pediatric Cardiomyopathies. Circ Res 2017;121:855-73. [Crossref] [PubMed]
- Augusto JB, Eiros R, Nakou E, et al. Dilated cardiomyopathy and arrhythmogenic left ventricular cardiomyopathy: a comprehensive genotype-imaging phenotype study. Eur Heart J Cardiovasc Imaging 2019. [Epub ahead of print]. [Crossref] [PubMed]
- Löwe T, Kley RA, van der Ven PF, et al. The pathomechanism of filaminopathy: altered biochemical properties explain the cellular phenotype of a protein aggregation myopathy. Hum Mol Genet 2007;16:1351-8. [Crossref] [PubMed]


