
A narrative review of precision medicine in neonatal sepsis: genetic and epigenetic factors associated with disease susceptibility
Introduction
Background
Neonatal sepsis is generally defined as a systematic condition caused by dysregulated host responses to infection, with pathogens ranging from bacteria to viruses and fungi (1-3). Based on the timing of onset, neonatal sepsis can be divided into either early-onset sepsis (EOS) or late-onset sepsis (LOS), with the classical cut-off point being the first 72 h after birth (4). In most cases, EOS results from vertical mother-to-infant transmission before and during delivery, whereas LOS is attributed to postnatal exposure to environmental organisms. Although overlap exists, the microorganisms isolated in EOS are typically different to those isolated in LOS. For example, Group B streptococcus and Escherichia coli are the top two causes of EOS, with infection rates of 43% and 29%, respectively (2), whereas coagulase-negative staphylococci and staphylococcus aureus are frequently detected in patients with LOS (5). Of note, considering the inherent vulnerability of neonates, opportunistic bacteria and fungi cannot be ignored. Neonatal sepsis may present asymptomatically or non-specifically with multiple systems involved. These symptoms can include temperature instability, poor feeding, respiratory distress, oliguria, diarrhea, jaundice, and purpura, which might mimic other diseases (3). The clinical usage of blood culture, the gold standard in diagnostics, is constrained by its low positive detection rate, long turnaround time and the influence of antibiotic administration (5). Due to its high negative predictive value and time- and sample-saving capabilities, quantitative real-time polymerase chain reaction (qPCR) of bacterial DNA is increasingly being used, but it is unable to conduct susceptibility tests or distinguish between active and resolved infections (6). Moreover, other culture-independent diagnostic tests, including complete blood count, C reactive protein, and procalcitonin, are regarded as being less than ideal indicators due to their lack of specificity (7).
Rationale and knowledge gap
Given the predicaments mentioned above, early diagnosis of neonatal sepsis is difficult; as a consequence, morbidity and mortality remain high despite advances in treatments. A more efficient prophylactic and diagnostic procedure is therefore urgently needed. The epidemiological study on twins suggests that hereditary factors act in conjunction with environmental factors to affect neonatal sepsis susceptibility (8). However, at present, little is known about the hereditary risks. Previous research has demonstrated that genomic polymorphisms change gene functions by altering transcription or translation, which is possibly involved in the pathophysiology of certain diseases including sepsis (9,10). Therefore, the identification of genetic components would potentially aid early risk stratification and individualized treatment and would provide the theoretical foundation for the clinical translation of precision medicine, which is a burgeoning paradigm intended to match the right interventions to the right population. To our knowledge, most existing papers are devoted to metabolomics, proteomics, or a subset of genomics (11-13), and no study to date has comprehensively summarized the hereditary risks of neonatal sepsis.
Objective
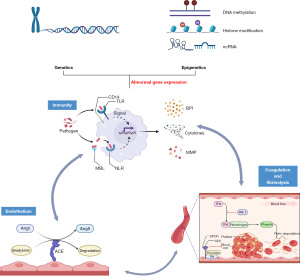
In this review, we will elaborate on the genetic and epigenetic factors of neonatal susceptibility to sepsis and endeavor to thoroughly define the genomic landscape underlying neonatal sepsis (Figure 1). We hope that integrating specific genomic makeup with other omics data will promote optimal precision medicine, where risk stratification, early diagnosis and personalized management are no longer dreams. We present the following article in accordance with the Narrative Review reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-22-369/rc).
Methods
A comprehensive search of PubMed was conducted using appropriate MeSH terms and keywords (Table 1). Without any restriction on the study type, articles published in English prior to June 1, 2022, were retrieved. Relevant pediatric, adult, and animal- and laboratory-based studies were reviewed wherever possible.
Table 1
| Items | Specification |
|---|---|
| Date of search | June 1, 2022 |
| Databases and other sources searched | PubMed |
| Search terms used | neonatal sepsis, pediatric sepsis, sepsis, genetics, epigenetics, polymorphism, variation, risk, susceptibility, precision medicine |
| Timeframe | Published before June 1, 2022 |
| Inclusion and exclusion criteria | There was no restriction to the study type but only articles in English were included |
| Selection process | The search was conducted by the first author using methods discussed and approved by both authors |
Genetic factors related to neonatal sepsis susceptibility
In sepsis, the immune and endothelium interact closely with the coagulation and fibrinolysis systems (1,9). Research indicates that functional changes of genes involved in these three systems, including substitutions, insertions, or deletions of base pairs, and copy number variants, are responsible for an individual’s predisposition to neonatal sepsis (14). The putative loci are discussed below and summarized in Table 2.
Table 2
| Gene name | Polymorphisms | Authors and publication year |
|---|---|---|
| TLR2 | rs5743708+2477G/A Arg753Gln | Kutukculer et al., 2007; Lu et al., 2019 |
| rs3804099+19216T/C Asn199Asn | Abu-Maziad et al., 2010; Martin et al., 2018 | |
| rs3804100T/C | Abu-Maziad et al., 2010; Martin et al., 2018 | |
| TLR4 | rs4986790+896A/G Asp299Gly | Ahrens et al., 2004; Härtel et al., 2007; Yuan et al., 2008; Sampath et al., 2013; Swierzko et al., 2016; Martin et al., 2018; Lu et al., 2019 |
| rs4986791+1196C/T Thr399Ile | Ahrens et al., 2004; Härtel et al., 2007; Yuan et al., 2008; Sampath et al., 2013; Swierzko et al., 2016; Martin et al., 2018; Lu et al., 2019 | |
| IRAK1 | rs1059703+1595T/C Leu532Ser | Sampath et al., 2013 |
| TOLLIP | rs5743867T/C | Fakhri et al., 2016 |
| NOD1(CARD4) | rs6958571 indel T/GG | Sampath et al., 2017 |
| NOD2(CARD15) | rs2066847+3020-/C Leu1007Pro | Ahrens et al., 2004; Härtel et al., 2007; Tekin et al., 2012 |
| rs2066844C/T Arg702Trp | Tekin et al., 2012 | |
| rs2066845G/C Gly908Arg | Tekin et al., 2012 | |
| CD14 | rs2569190-159G/A (-260C/T) | Ahrens et al., 2004; Baier et al., 2006; Abu-Maziad et al., 2010; Zhang et al., 2013; Esposito et al., 2014; Martin et al., 2018; Mustarium et al., 2019 |
| MBL2 | A/O haplotype (rs5030737+154C/T Asp52Cys, rs1800450+161G/A Gly54Asp, rs1800451+170G/A Gly57Glu | Ahrens et al., 2004; Frakking et al., 2007; Härtel et al., 2007; Dzwonek et al., 2008; van et al., 2008; Abu-Maziad et al., 2010; Koroglu et al., 2010; Cinzia et al., 2010; Ozkan et al., 2012; Luo et al., 2014; Swierzko et al., 2016; Badawy et al., 2018; Martin et al., 2018; Lu et al., 2019 |
| TNFα | rs1800629-308G/A | Hedberg et al., 2004; Sipahi et al., 2006; Härtel et al., 2011; Srinivasan et al., 2017 |
| IL-1B | rs16944-511G/A | Abu-Maziad et al., 2010; Esposito et al., 2014; Allam et al., 2015; Mustarium et al., 2019; Varljen et al., 2020 |
| rs143634+3954C/T | Balding et al., 2003; Treszl et al., 2003; Abu-Maziad et al., 2010; Zhang et al., 2014; Lu et al., 2019 | |
| IL-6 | rs1800795-174G/C | Kilpinen et al., 2001; Balding et al., 2003; Treszl et al., 2003; Harding et al., 2003; Ahrens et al., 2004; Gopel et al., 2006; Sipahi et al., 2006; Michalek et al., 2007; Reiman et al., 2008; Abu-Maziad et al., 2010; Zidan et al., 2014; Allam et al., 2015; Gao et al., 2015; Srinivasan et al., 2017 |
| IL-8(CXCL8) | rs4073-251T/A | Abu-Maziad et al., 2010; Esposito et al., 2014; Hu et al., 2016; Fu et al., 2019; Lu et al., 2019 |
| IL-10 | rs1800896-1082A/G | Balding et al., 2003; Treszl et al., 2003; Baier et al., 2006; Abu-Maziad et al., 2010; Emonts et al., 2010; Esposito et al., 2014; Pan et al., 2015; Srinivasan et al., 2017 |
| BPI | rs4358188+645A/G Lys216Glu | Michalek et al., 2007; Abu-Maziad et al., 2010; Esposito et al., 2014; Martin et al., 2018; Mustarium et al., 2019 |
| rs5743507+545G/C Val182Val | Michalek et al., 2007 | |
| MMP16 | rs2664349+39811 A/G | Esposito et al., 2014; Mustarium et al., 2019 |
| ACE | rs4646994/rs4340 intron16 289bp Alu ins/del | John et al., 2005; Cogulu et al., 2008; Spiegler et al., 2010; Hou et al., 2015; Dou et al., 2017; Lu et al., 2019; Jarahzadeh et al., 2022 |
| Factor V | rs6025+1691G/A Arg506Gln | Härtel et al., 2006 |
| Factor II | 20210G/A | Härtel et al., 2006 |
| Factor VII | 323del/ins | Härtel et al., 2006 |
| Factor XIII | rs5985C/A Val34Leu | Härtel et al., 2006 |
| PAI-1 | rs1799768-675 5G/4G | Li et al., 2013; Shi C et al., 2015; Shi Q et al., 2015; Lu et al., 2019; Jarahzadeh et al., 2022 |
| EPCR | exon3 23bp del/ins | Taylor et al., 2000; Esmon et al., 2003; Sipahi et al., 2006 |
| VDR | rs2228570T/C (FokI) | Das et al., 2016; Zeljic et al., 2017; Tayel et al., 2018 |
| rs731236C/T (TaqI) | Das et al., 2016; Zeljic et al., 2017; Tayel et al., 2018 | |
| rs2107301 | He et al., 2021 | |
| rs2189480 | He et al., 2021 | |
| rs9729 | He et al., 2021 | |
| rs2239815 | He et al., 2021 | |
| rs3782905 | He et al., 2021 | |
| rs4516035T/C | He et al., 2021 | |
| rs7139166 | He et al., 2021 | |
| rs11168266 | He et al., 2021 | |
| rs11168293 | He et al., 2021 | |
| rs739837 | Xiao et al., 2022 | |
| TREM1 | rs2234246G/A | Xiao et al., 2022 |
TLR, Toll-like receptor; IRAK1, interleukin-1R-associated protein kinase 1; TOLLIP, Toll-interacting protein; NOD, nucleotide-binding oligomerization domains; CD14, cluster of differentiation 14; MBL, mannose-binding lectin; TNFα, tumor necrosis factor alpha; IL, interleukin; BPI, bactericidal permeability increasing protein; MMP, matrix metalloproteinase; ACE, angiotensin converting enzyme; PAI-1, plasminogen activator inhibitor 1; EPCR, endothelial protein C receptor; VDR, vitamin D receptor; TREM1, triggering receptor expressed on myeloid cells 1.
Genes involved in the immune system
Recognition of pathogenic components
Toll-like receptors (TLRs) and relevant signaling molecules
TLRs, which comprise dozens of members, are a battery of receptors expressed in immune cells. They participate in a critical mechanism in recognizing various patterns of invading pathogens and subsequently inducing signaling transduction pathways (15,16). In recent decades, increasing evidence has shown whether polymorphisms in genes coding for or in the regulation of these pleiotropic receptors can affect the neonates’ hereditary propensity to sepsis (17,18).
For TLR2, rs5743708G/A Arg753Gln has been investigated with mixed findings. In 2007, a case-control study of 143 Turkish children indicated a close relationship between the Arg753Gln polymorphism and a tendency for recurrent febrile bacteremia (19). Intriguingly, a recently published meta-analysis implied that unlike adults, children were marginally protected from sepsis by this genetic alteration (20). Another two polymorphisms, rs3804099T/C and rs3804100T/C, were found to be characterized by a potent correlation with neonatal sepsis in a retrospective study (21); however, a later investigation failed to produce similar results (18).
Regarding TLR4 rs4986790A/G Asp299Gly and rs4986791C/T Thr399Ile, two of the most frequent hotspots, Sampath et al. carried out a prospective nested case-control study of 408 neonates with very low birth weight (VLBW, birth weight <1,500 g) and reported that both rendered neonates susceptible to gram-negative infections even in regression models with control for confounders (P=0.004) (22), which is consistent with the findings from Yuan et al. (23). Conversely, several other relevant investigations found that neither rs4986790 nor rs4986791 was linked with neonatal susceptibility to sepsis (18,20,24-26).
Molecules that interact with TLRs and then modulate downstream signals, such as interleukin-1 receptor-associated protein kinase 1 (IRAK1) and toll-interacting protein (TOLLIP), are also of clinical relevance. Sampath et al. reported that female infants with VLBW who had the IRAK1 rs1059703C/T genotype tended to experience fewer gram-negative infections (22), and TOLLIP rs5743867T/C was found to decrease the sepsis risk in patients undergoing complex open-heart surgery who were under 1 year of age (27). Furthermore, additional variants have been topics of interest and studied in adults (28). Given that age is considered to be a confounder, more confirmatory research is needed to clarify whether these candidate variations play a pivotal role in neonates.
Nucleotide-binding oligomerization domains (NOD)-like receptors (NLRs)
NLRs serve as the receptors to diverse components derived from invasive microorganisms and induce autophagy for antigen presentation (29,30). NOD1 and NOD2, two members of the NLR superfamily, have attracted considerable attention. In an international cohort, Sampath et al. verified that the C allele of NOD1 rs6958571 significantly increased the prevalence of gram-positive bacterial bloodstream infection in extremely low birth weight (birth weight <1,000 g) and Caucasian preterm neonates (31). In a study of 356 infants, Härtel et al. noted that those with VLBW and NOD2-3020insC mutation were more likely to suffer from blood culture-proven sepsis (25); however, Ahrens et al. drew a different conclusion (24). Additionally, Tekin et al. evaluated three common mutations within the NOD2 gene (Arg702Trp, Gly908Arg, and Leu1007Pro) and illustrated their contributions to septic susceptibility, which highlighted the role of NOD2 (32).
Cluster of differentiation 14 (CD14)
CD14 is an adjuvant co-receptor with a constitutive expression in a large variety of cells, which aids better recognition of pathogenic components by TLRs (33,34). CD14 rs2569190-159G/A, which is identical to -260C/T, has been explored in newborns with conflicting results. A retrospective study recruiting 293 mechanically ventilated infants with VLBW showed effects of the T allele on multiple episodes of blood stream infection, of which the strongest was in African-American infants (CC: 15%, CT: 11%, TT: 39%, P=0.003) (35). Another survey seems appeared to verify this tendency, where Esposito et al. considered this polymorphic site to be a risk factor for severe sepsis (36). Nonetheless, Martin et al. argued a protective effect of the T/T genotype on neonatal sepsis (18), but several other meta-analyses were unable to demonstrate any association (21,24,37,38). Inherent differences, such as ethnicity and comorbidities, may in part explain these divergent results.
Mannose-binding lectin (MBL)
MBL, a type of opsonin, plays a part in the binding of pathogenic components, boosting of lectin-dependent complement activation, and mediation of phagocytic clearance of pathogens (39-41). MBL2 rs5030737, rs1800450, and rs1800451 form three distinct alleles, named B, C, and D, respectively, and are collectively known as O, with A being the wild type. Cumulative evidence confirmed that a low serum MBL concentration resulting from O was associated with a higher incidence of sepsis in all age groups (20,26,42-46). Unfortunately, this theory did not hold in observations focused on neonatal sepsis, in which no such correlation was found (18,21,24,25,47-49). The mixed data mentioned above raise the question as to the exact role of MBL2 in neonatal sepsis.
Cytokine responses
Tumor necrosis factor alpha (TNFα)
Soon after an infectious stimulus, a host with normal immune function is flooded with a crucial cytokine TNFα, triggering a succession of primitive responses to invasion (50). In this regard, aberrance in the quality or quantity of TNFα secretion caused by polymorphic variations might remodel the susceptibility to, or prognosis of, sepsis. Among several candidate mutations, TNFα rs1800629-308G/A stands out as having high potential. Through in vitro experiments, the rarer A allele was delineated to increase the transcriptional and translational activities of TNFα compared with the G allele (51). Sipahi et al. evaluated 53 children with severe sepsis aged 0 to 15 years and found that allele A-carrying status was related to the prevalence but not the outcome (52). In a retrospective cohort with 173 mechanically ventilated infants with VLBW, the allele A was reported to be a genetic modulator of mortality once sepsis had developed (53). However, these findings could not be replicated in the following research (54,55). Moreover, a pattern of predisposition conferred by the G allele was observed in a group of Saudi term neonates (56), which concurred with the findings of an updated meta-analysis on the pediatric subgroup (57). Even though these discoveries have linked -308G/A with the predisposition to and the severity of sepsis, an explicit conclusion cannot be drawn due to the lack of studies in larger cohorts and with more stringent control for confounders.
Interleukin (IL)-1
IL-1 is a potent member of the chemokine superfamily and is generally accepted as an integral mediator in the pathogenesis of sepsis. Three disparate polypeptides—IL-1α, IL-1β, and IL-1Ra (IL-1 receptor antagonist) constitute the whole IL-1 family (58). As early as 1993, elevated IL-1β was detected in neonates with EOS compared with controls (59), which provided a rationale for speculation that genetic variability fluctuating IL-1β level might modulate the course of neonatal sepsis.
Regarding IL-1B, the coding gene of IL-1β, Varljen et al. discovered an enrichment of the rs16944-511AA genotype in preterm infants suffering from and succumbing to EOS (60). Although no positive association was reported by Abu-Maziad et al. (21), Esposito et al. ascertained a notable increase in the overall risk of sepsis development in preterm infants with the rs1143643 TT or CT genotype, which supported the latter observation (36). However, in a Saudi Arabian neonatal cohort, CC genotype carriers presented with a higher concentration of circulating IL-1β and an increased risk of EOS. IL-1B rs143634+3954C/T is a synonymous variant with no amino acid changes, and its TT genotype was linked with increased IL-1β but decreased susceptibility to sepsis in adults (20,61); however, similar conclusions have not been drawn in younger individuals so far (21,62,63). It is worth pointing out that such discernable age gap awaits to be addressed, and these results will add evidence to the notion that the balance of the ratio of IL-1 to IL-1Ra plays a role in sepsis progression, rather than IL-1 or IL-1Ra alone.
IL-6
IL-6 is a proinflammatory cytokine that activates lymphocytes, induces pyrogen, and accelerates acute-phase reactants and antibody synthesis (64). These pleiotropic properties make IL-6 an integrative index and a biologically plausible candidate gene of sepsis. A pilot study involving neonates illustrated that lipopolysaccharide induced higher IL-6 expression in monocytes isolated from carriers of the C allele of rs1800795 (65), and this finding was supported by several subsequent articles in which the C allele was concluded to be a sepsis-predisposing and/or -modifying variant (35,66,67). Discrepant results are unsurprising considering of the diversity in research methods and the general characteristics of samples. Harding et al. put forward that the G allele might impair the defense against bacteria (68), and Zidan et al. verified an association of the G allele with community-acquired pneumonia (69). Similarly, increased EOS frequency, repeated septicemia episodes, and decreased survival rate were reported in GG homozygotes (24,56,62). Beyond that, several studies failed to obtain any correlation (21,52,55,63,70,71). Excessive systemic release of IL-6 is thought to be a core in the pathophysiology of sepsis, but which allele is responsible has yet to be determined.
IL-8
IL-8, also termed CXCL8, possesses the ability to motivate neutrophil recruitment, adjust inflammatory reactions, and promote angiogenesis (64). Studies have observed that IL-8 rs4073-251T/A boosted IL-8 production under lipopolysaccharide stimulation and, thus, exerted an influence on various pathological conditions to a certain degree (72-75). Hu et al. found that males carrying the T allele or TT genotype were more susceptible to sepsis (76), which was diametrically opposite to Lu et al. and Fu et al. (20,77). Interestingly, focusing on premature infants, Esposito et al. concluded that the AT genotype aggravated the development of sepsis (36), whereas Abu-Maziad et al. dismissed any effect caused by this mutation (21). Admixture of ethnicity and sex, combined with the distinct definitions used, might partially explain why such notable conflicts have emerged.
IL-10
Under normal circumstances, to maintain homeostasis, overwhelming proinflammatory responses are under tight regulation by anti-inflammatory mediators, of which IL-10 works best. IL-10 rs1800896-1082A/G is positioned in the 5'-flanking region with higher IL-10 inducibility (78). Similar to Baier et al. and Balding et al. (35,62), Srinivasan et al. clarified a defensive function of the GG genotype (odds ratio =0.51) (55). However, not all studies have obtained statistically significant results (36,63,79). Abu-Maziad et al. illustrated an inverse relationship between the GG genotype and gram-negative infection risk in a large preterm cohort (21), which corresponded with results from a meta-analysis confirmed in an Asian population (80). The insufficient reproducibility may be explained by developmental differences; that is to say, the propensity or progression of sepsis is influenced by the degree of immune system maturation.
Other immunological genes
Bactericidal permeability increasing protein (BPI)
With high potency and affinity to lipopolysaccharide, BPI acts as a key effector in antibacterial defense. The increased risk of gram-negative bacterial infection can be partially attributed to selective BPI deficiency in neonates (81), and a randomized controlled trial proposed that recombinant BPI held promise as an adjunctive treatment for children with severe meningococcal sepsis (82). According to Esposito et al., the rs4358188 AG genotype protected premature neonates from sepsis (36), and Abu-Maziad et al., Martin et al., and Mustarim et al. did not detect any genetic risk profile of BPI alleles (18,21,37). Moreover, Michalek et al. noted that children carrying rs4358188 GG combined with rs5743507 AG or GG haplotypes were more likely to suffer from gram-negative sepsis and resultant complications (83).
Matrix metalloproteinases (MMPs)
Several lines of evidence have made it clear that zinc-relevant MMPs are not only matrix-degrading enzymes but also function as a mechanism in immune modulation (84,85). While appropriate MMP secretion accelerates infection eradication, excess MMP production becomes a tissue destroyer, which favors microorganism dissemination and persistence. This lays the foundation for MMP genetic polymorphisms being a logical candidate (85). MMP-16 is a new player in bronchopulmonary dysplasia (86), whose rs2664349+39811 GG genotype was described as being closely correlated with bacteriologically proven sepsis (36). Findings from a cross-sectional study suggested that this G to A mutation might eventually pose a danger, as most preterm infants in both the proven and unproven neonatal sepsis groups exhibited variations, albeit with inadequate statistical significance (37). As a corollary, preparations targeting aberrant MMP have been evaluated for the adjunctive treatment of patients with sepsis and septic shock including retinoids and glucocorticoids (2).
Genes involved in endothelial factors
Chiefly distributed on epithelial cells, the carboxypeptidase angiotensin converting enzyme (ACE) functions as a crucial regulator of hemodynamic stasis by converting angiotensin (Ang) I to Ang II and participating in bradykinin catabolism (87). In both in vivo and in vitro studies, ACE deficiency has been widely characterized in patients suffering from sepsis, indicating a potential role of ACE in sepsis (87,88). The variation consisting of either the presence (insertion, I) or the absence (deletion, D) of a 287 bp Alu repeat fragment has been assessed and is believed to be a leading contributor in determining serum ACE concentration (89). Findings on the predictive value of allele D in the propensity to and the outcome of sepsis are extremely inconsistent in different studies (20,90,91). In keeping with Spiegler et al. (92), John Baier et al. discovered comparable rates of sepsis mortality and morbidity independent of ACE I/D genotype in mechanically ventilated infants with VLBW (93). On the other hand, a retrospective study demonstrated an adverse effect of I allele on pediatric sepsis development (94), providing favorable evidence for a subsequent meta-analysis (91). However, this was conclusively proven to be incorrect by a more recent one (95). Selection bias should be taken into consideration since mechanical ventilation and low birth weight can predispose newborns to sepsis (2).
As a mandatory participant of the renin-angiotensin-aldosterone system (RAAS), there is not only an ongoing debate regarding the influence of ACE I/D on sepsis risk and outcomes but also in relation to the therapeutic value of RAAS derivatives (96-99). Despite the current lack of an unequivocal answer, in no way can we exclude the possibility of other variations or haplotypes of the ACE coding gene or linkage disequilibrium with functional genetic determinants.
Genes involved in the coagulation and fibrinolysis system
Homeostasis is badly disrupted during sepsis and a vicious circle occurs. Fulminant inflammation elicited by cytokines or other proinflammatory elements concurrently exacerbates the coagulation cascade, which in turn amplifies inflammation (100). This mutual independence underlines the notion that malfunction in any part of the coagulation pathway may modify the onset and evolution of sepsis and, consequently, offer novel insights into biomarkers and therapeutic implications for clinical exploration, especially in naturally hypercoagulable newborns (101,102).
Coagulation factors
Culminating in thrombin generation, the intrinsic and extrinsic coagulation pathways are regulated by 13 coagulation factors (Factor I to XIII). Härtel et al. removed Factor V rs6025G/A Arg506Gln (Factor V Leidon or FVL), Factor II 20210G/A, and Factor VII 323del/ins from the list of candidate inherited risk predictors, and simultaneously illustrated that the rs5985C/A Val34Leu mutation of Factor XIII—a transglutaminase that enhances the stability of cross-linked fibrin polymers through bridging bonds between monomers—predisposed carriers to neonatal sepsis (103). It is tempting to postulate that transformation of the fibrin meshwork to premature structures with thinner fibers and smaller pores as well as the resultant less vigorous response prompted by Val34Leu may contribute to risk alterations (104,105), but a tenable interpretation is currently lacking due to shortcomings in reproducibility.
Plasminogen activator inhibitor-1 (PAI-1)
Fibrin is viewed as a double-edged sword in that moderate generation prevents bleeding while overzealous deposition gives rise to vessel occlusion and, in severe cases, organ dysfunction. Degradation of fibrin is mostly performed by plasmin, a ubiquitous proteinase existing in the form of deactivated plasminogen. Plasminogen activator converts the precursor to its active form, whose proteolytic contributions can be rapidly diminished by specific blocker PAI-1. The PAI-1 gene possesses an insertion/deletion of G residue polymorphism (either the 4G or 5G allele), of which the 5G allele accounts for profoundly decreased plasma PAI-1 concentration secondary to repressed transcription (106). Shi et al. suggested no association between PAI-1 4G/5G with sepsis (107), which was dramatically overturned in the same year (108). Recently, Jarahzadeh et al. published literature restricted to pediatric cases, and they stated that children with the 4G allele showed higher incidence of sepsis than their peers (95), which was identical to the conclusions of Lu et al. and Li et al. (20,109). Case-control reports involving neonates leave much to be desired, and caution is warranted until rigorous experimental verification becomes available.
Endothelial protein C receptor (EPCR)
EPCR is a receptor that indirectly facilitates PC activation by presenting PC to the membrane-bound thrombin-thrombomodulin complex, and exhibits an anti-inflammatory function in animal experiments (110,111). Sipahi et al. once explored a 23 bp insertion mutation within exon 3 of the EPCR gene and found a significantly higher odds ratio in children with sepsis (52). Together with other supporting evidence (112,113), these emerging findings pave the way to a better understanding of neonatal sepsis.
The demise of recombinant human activated PC marks the last chapter of the story and addresses a conundrum about the real role of coagulation in sepsis (114). As Fiusa et al. discussed from an evolutionary medicine perspective, a threshold might exist, above which coagulation activation is harmful and below which it enhances pathogen clearance (115). This again emphasizes the extreme complexity of the sepsis triad of inflammation, coagulation, and fibrinolysis, and more importantly, provides a unique insight into sepsis pathophysiology and tailored interventions.
Other potential genes
Vitamin D receptor (VDR)
In addition to being a classical modulator of calcium and phosphorus homeostasis, vitamin D exhibits antimicrobial and anti-inflammatory properties in a VDR-binding method (116,117). Emerging evidence has revealed a trend of alteration in vitamin D production and stability driven by several VDR gene polymorphic differences (118). For example, assessing rs2228570T/C (FokI) and rs731236C/T (TaqI) in a small Indian cohort, Das et al. discovered vitamin D insufficiency in participants with sepsis, but the distributions of both FokI and TaqI deviated from the Hardy-Weinberg equilibrium (119). TaqI has not been indicated to have any association in subsequent reports (120,121). Regarding the silent mutation FokI, the TT genotype and T allele were delineated as a hazard to vitamin D expression and EOS development (121), which was the exact opposite of findings in Serbian adults (120). Recent efforts involving other allelic loci have further underscored the association between the VDR gene and sepsis (122). Of note, other than the G allele of VDR rs739837, Xiao et al. stated that the rs2234246 T allele of triggering receptor expressed on myeloid cells 1 (TREM-1), an immunoglobulin activated transmembrane receptor, could predict neonatal sepsis (123). One can easily posit that vitamin D supplements can have a favorable effect in sepsis prevention and alleviation, but whether VDR polymorphisms and haplotypes are eligible to be included in the expanding array of genetic markers remains to be established.
On balance, whether and how genetic alterations of this “restless warrior” contribute to individuals’ variability in sepsis have yet to be explored. The current evidence represents only a small step in the assessment of the contribution of genetics to sepsis, and studies with high homogeneity are needed to correlate existing single nucleotide polymorphisms with specific products and other separate causative polymorphisms based on linkage disequilibrium.
Epigenetic factors related to neonatal sepsis susceptibility
Epigenetic traits emerge as a new layer of reshaping genomic topology without alterations in the DNA sequence. This fine-tuned heritable phenomenon, by and large, encompasses DNA methylation, non-coding RNAs (ncRNAs), and post-transcriptional modifications of histones, and functions as a gene-specific ‘volume control’ to activate or silence gene transcription (124,125). Evidence has shown that epigenetic changes participate in short- and long-term events of sepsis (126,127). Below, we outline a framework of recently proposed epigenetic signatures concerning sepsis.
Histones, a group of proteins responsible for packaging lengthy DNA strands into highly organized chromatin and post-transcriptional modifications, have a share in modifying chromatin dynamics and gene transcription. Underscoring the finding of sepsis-induced alterations in histone post-transcriptional modifications (128), Bermick et al. delineated a unique H3K4me3 pattern in neonatal monocytes exposed to chorioamnionitis (129). In synergy with changes in several known immune genes, this H3K4me3 pattern resulted in an insufficient defense against secondary microbial challenge, predisposing neonates to sepsis attack (129). Structural maintenance of chromosome (Smc) 4, a core subunit of condensin, was found to promote NF-κB essential modulator transcription to augment TLR- and virus-triggered proinflammatory responses by recruiting H4K5 acetylation (130). Furthermore, Smc4 knockdown protected mice from sepsis and sepsis-related mortality (130). These observations provided a basis for future explorations in risk prediction and even gene-editing therapy for neonatal sepsis.
DNA methylation is a chemical process where human DNA methyltransferases add a methyl group to cytosine, predominantly at CpG dinucleotides (131). In line with assessments in adults (132,133), differential methylation profiles were disclosed in neonates with and without sepsis, suggesting a potential use of DNA methylation as an indicator for neonatal sepsis prediction and subtype distinction (134). A similar result was yielded by a small epigenome-wide association study in which up to 81 differentially methylated CpGs mapped in 64 genes emerged with biological and clinical relevance (135). For example, protocadherin beta (PCDHB) genes, a panel of genes involved in calcium-dependent cell adhesion and antigen presentation, were hypermethylated in neonates with sepsis compared with healthy controls (135). According to Tendl et al., pathogen-specific DNA methylation changes were only detected in the promoter region of the procalcitonin-relevant calcitonin-related polypeptide α (CALCA) gene and not in other simultaneously activated genes (TLR4, MyD88, and CRP) during neonatal sepsis (136). These findings mean that PCDHBm and CALCAm, along with other rarely studied sites, should be included into the list of epigenomic biomarkers of neonatal sepsis.
ncRNAs comprise a different set of molecules which are not translated to functional proteins (137). Aberrant expression profiles of ncRNAs have been consistently detected in various cell types and tissues from individuals with sepsis (138-140). Additionally, using samples from 87 neonates with sepsis or respiratory infection, Wang et al. evaluated the levels of miRNAs, which were considered to be indicators of adult sepsis (141). Up-regulation of miR-15a/16 was observed in cases with sepsis, the molecular mechanism of which pointed to the suppression of lipopolysaccharide-promoted TLR4/IRAK1 signaling (141). Another study of neonatal sepsis identified 59 differentially expressed miRNAs between the case and control groups (including miR-181a, miR-141, and miR-143), and miR-16 was found to be down-regulated in gram-negative sepsis (142). Although these data suggest that some ncRNAs hold potential as potent fingerprints in neonatal sepsis, no consensus has been reached as to which single ncRNA or ncRNA panel bears maximal sensitivity and specificity while being translated into clinical use.
Overall, the data mentioned above theoretically reinforce the developmental programming hypothesis that, as an interface between the genome and the environment, epigenetic marks are rewritten in response to the early environmental input, maintaining a perpetual memory of alteration and holding the potential to provoke susceptibility to specific diseases in later life (143). There are promising signs that the development of reliable epigenetic biomarkers based on personal profiles for sepsis risk prediction, proper management, and prognostic assessment is on the horizon.
Precision medicine in neonatal sepsis
Timely and accurate diagnosis based solely on clinical and laboratory parameters has achieved only partial success. As a result, there is a relatively low threshold for clinical suspicion of infection and extensive empirical antibiotics usage due to concerns about catastrophic collapse secondary to delayed management. Furthermore, from the latter arises another concern about drug resistance. Undertreatment or overtreatment poses a profound threat to neonatal health, and continuous efforts have been dedicated to the search for an appropriate solution. The evidence mentioned above lays a theoretical foundation for the clinical translation of precision medicine, which is a burgeoning paradigm proposed to match the right interventions to the right population.
Polygenic risk scores (PRSs) have recently made strides in providing risk discrimination in several heterogenous clinical entities, including breast cancer and type 2 diabetes (144,145). This quantitative metric summarizes individual hereditary liability to a disease or a trait by calculating the cumulative impact of a range of genetic polymorphisms according to their respective weighed effect sizes (146). For sepsis, a single variant inadequately reflects the overall risk, but partial susceptibility may be captured when pooling a suite of genetic alleles, rendering the application of PRSs possible. Lu et al. extracted 17 variants from 64 candidate loci to construct a weighted genetic risk score, which was confirmed to be positively correlated with traumatic sepsis morbidity and fitted better when the injury severity score was added (147). Similarly, another PRSs program showed a good performance in screening out high-risk individuals (148). The PRSs of patients with septic shock reflected clinically relevant traits at genomic level, with the PRSs for a higher level of CRP being related to a higher septic shock risk (149). These interesting findings inform the translation from the percentile of PRSs to a quantitative estimation of an individual’s predisposition to sepsis, which not only allows for timely prophylactic methods such as pathogen exposure attenuation and antibiotic interventions direct for those at-risk, but avoids unnecessary waste of resources. Despite the existing pioneering work, the PRSs have only just started to be translated from the bench to the bedside, and trials specific to neonates have yet to be conducted.
Evidence is accumulating that some of the abovementioned genetic variants are involved at different points of the disease trajectory. For instance, TNFα rs1800629-308G/A was designated as a popular genetic biomarker of susceptibility to and severity of sepsis in newborns (53,56), a probabilistic explanation of which might be the resultant excessive TNFα production and consequent uncontrolled tissue damage. An innovative macrophage-targeted RNA-interference system achieved the goal of down-regulation of TNFα, indicating a fundamental therapy for sepsis free of side effects, and analogous indications could be extrapolated to other polymorphisms (150). Moreover, emerging themes such as gene therapy and genetic editing systems hold promise to be included as adjunctive approaches once an in-depth complete genomic map has been drawn (151).
With the advent of the omics-data-driven era, identifying a genetic background at birth has become a feasible tool for prognostic and predictive enrichment and stratify neonates into cohorts with more homogeneity (12). Such precise stratification based on interrogation of genome will evolve to provide the basis for tailored managements, which is derived from the underpinning molecular mechanisms rather than common phenotypic signs. Nonetheless, given the unique predicaments in neonatology, there is still a long way to go before precision medicine can be fully embraced in the field of neonatal sepsis.
Discussion
Despite the progress in multidisciplinary treatment, a growing number of neonates are confronted with the potentially fatal challenge of sepsis after birth, which remains difficult to detect in its early stage. The epidemiological study on twins suggests that hereditary factors together with environmental factors modify the risk of neonatal sepsis (8). Understanding hereditary susceptibility comprehensively is therefore a cornerstone in further optimizing the risk assessment, continuous monitoring, accurate diagnosis, and personalized management of this vulnerable population.
In this review, the pathophysiological process of neonatal sepsis was divided into the following three parts and the genetic factors were summarized: the immune, endothelium, and coagulation and fibrinolysis systems. A single locus is unlikely to account fully for susceptibility to sepsis. Instead, it is the synthetic action of multiple causal loci, each of which has a slight effect, combined with other environmental risk factors, that determines the ultimate sepsis phenotype. Inconsistent results point to a flip-flop phenomenon, which describes opposite contributions made by the same allelic variant within the same disease based on different studies (152). Such discrepancies regarding the impact of a certain genetic polymorphism on susceptibility to neonatal sepsis may be explained by the following reasons to some extent. Firstly, the conspicuous heterogeneity of study population covers gestational age, birthweight, and comorbidities, which may mask the slight effect conferred by risk alleles. Secondly, some studies show methodological weaknesses in the selection of participants, where the controls might not have exposed to similar pathogens as cases. Therefore, more appropriate inclusion criteria are warranted, such as enrolling well-matched infectious individuals without deteriorating into sepsis. Thirdly, false positives or false negatives could be attributed to the relatively small sample sizes of studies. Fourthly, as allele frequencies, linkage disequilibrium patterns, and effect sizes of common polymorphisms vary with ancestry, caution should be exercised when extrapolating results to populations with different ancestral backgrounds. The fifth explanation relates to discrepancies in clinical phenotypes. It is problematic to include both EOS and LOS in terms of pathogenic microorganisms and immune maturation. Moreover, a consensus on neonatal sepsis definition is essential for the interpretation of blended findings. Finally, a polymorphism which has been demonstrated to be correlated with the risk or severity of sepsis, may, in reality, not be directly but rather indirectly involved through linkage with the actual genetic variation. Given that genes are not isolated, linkage disequilibrium, extended haplotype of a set of allelic mutations, and gene-environment interplay are issues that are too crucial to be dismissed without closer consideration.
Potential epigenetic mechanisms are discussed in the light of histone modifications, DNA methylation, and ncRNAs. The causal relationship between the two elements—that is, whether the pathogen exposure drives changes in epigenetics, or epigenetic signatures initiate development of sepsis—remains to be clarified. However, as James Watson said that “You can inherit something beyond the DNA sequence. That’s where the real excitement of genetics is now.” (153), epigenetics is still a frontier in biology in terms of understanding the possible mechanisms underpinning neonatal sepsis and delineating the new paradigm of precision medicine.
Finally, we introduce the prospect of applying precision medicine philosophy to neonatal sepsis. The inherent nature of neonatal medicine that constant vigilance and monitoring posed on babies are usually over protracted periods from birth, renders it an ideal setting for moving precision medicine to the clinical arena. The drawbacks of our current approach appear to be offset by the superiority of PRSs in remaining stable throughout life and enabling predictions to be made before noticeable symptoms. Moreover, gene therapy and immunomodulators based on individual genomic architecture might offer adjunctive resolutions to the emergence of antimicrobial-resistant bacteria. However, there remain some obstacles to transforming genomic data into practical paradigms, such as the cost-effect ratio, the training of clinicians, access to ancestry-matched gene banks, and, most importantly, negative public attitudes toward gene testing. Although skepticism regarding the adoption of precision medicine remains, a wealth of omics data and bioinformatics tools will assist with the implementation of precision medicine.
Strengths and limitations
To best of our knowledge, this is the first review to summarize the genetic and epigenetic risks of neonatal sepsis in detail and to outline promising opportunities for precision medicine. However, we must acknowledge, as a limitation of our review, that only published English language sources were retrieved. Furthermore, the drawing of stronger conclusions warrants studies with improved methodology.
Conclusions
To conclude, inherited susceptibility to neonatal sepsis has been reviewed in detail in terms of genetics and epigenetics, which has shown that the candidate factors, together with pathways they participate in, play key roles in modifying host-pathogen interaction. Although controversies exist, it is time to integrate genomic sequencing into routine protocols for neonatal sepsis. In the future, precision medicine will make risk stratification, early diagnosis, and personalized management achievable. Future efforts are warranted to capture the full spectrum of inherited susceptibility to neonatal sepsis, focusing on the collection of additional sources of evidence such as gene knockout experiments, the development of functional machine-learning approaches, and the construction of global-wide biobanks. Furthermore, the requirements for legal and ethical frameworks regarding patient privacy should always be kept in mind to protect against genetic discrimination.
Acknowledgments
We extend our sincere gratitude to Bin Chen and Youli Chen for their help in polishing our paper.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-22-369/rc
Conflicts of Interest: Both authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-22-369/coif). The authors have no conflicts of interest to declare.
Ethical Statement:
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Singer M, Deutschman CS, Seymour CW, et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016;315:801-10. [Crossref] [PubMed]
- Shane AL, Sánchez PJ, Stoll BJ. Neonatal sepsis. Lancet 2017;390:1770-80. [Crossref] [PubMed]
- Odabasi IO, Bulbul A. Neonatal Sepsis. Sisli Etfal Hastan Tip Bul 2020;54:142-58. [PubMed]
- Wynn JL, Wong HR, Shanley TP, et al. Time for a neonatal-specific consensus definition for sepsis. Pediatr Crit Care Med 2014;15:523-8. [Crossref] [PubMed]
- Kim F, Polin RA, Hooven TA. Neonatal sepsis. BMJ 2020;371:m3672. [Crossref] [PubMed]
- Oeser C, Pond M, Butcher P, et al. PCR for the detection of pathogens in neonatal early onset sepsis. PLoS One 2020;15:e0226817. [Crossref] [PubMed]
- Cantey JB, Lee JH. Biomarkers for the Diagnosis of Neonatal Sepsis. Clin Perinatol 2021;48:215-27. [Crossref] [PubMed]
- Bizzarro MJ, Jiang Y, Hussain N, et al. The impact of environmental and genetic factors on neonatal late-onset sepsis. J Pediatr 2011;158:234-8.e1. [Crossref] [PubMed]
- Gotts JE, Matthay MA. Sepsis: pathophysiology and clinical management. BMJ 2016;353:i1585. [Crossref] [PubMed]
- Hill AV. The immunogenetics of human infectious diseases. Annu Rev Immunol 1998;16:593-617. [Crossref] [PubMed]
- Bardanzellu F, Fanos V. How could metabolomics change pediatric health? Ital J Pediatr 2020;46:37. [Crossref] [PubMed]
- Ng S, Strunk T, Jiang P, et al. Precision Medicine for Neonatal Sepsis. Front Mol Biosci 2018;5:70. [Crossref] [PubMed]
- Keij FM, Achten NB, Tramper-Stranders GA, et al. Stratified Management for Bacterial Infections in Late Preterm and Term Neonates: Current Strategies and Future Opportunities Toward Precision Medicine. Front Pediatr 2021;9:590969. [Crossref] [PubMed]
- Srinivasan L, Kirpalani H, Cotten CM. Elucidating the role of genomics in neonatal sepsis. Semin Perinatol 2015;39:611-6. [Crossref] [PubMed]
- Akira S, Takeda K, Kaisho T. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat Immunol 2001;2:675-80. [Crossref] [PubMed]
- Leulier F, Lemaitre B. Toll-like receptors--taking an evolutionary approach. Nat Rev Genet 2008;9:165-78. [Crossref] [PubMed]
- Schröder NW, Schumann RR. Single nucleotide polymorphisms of Toll-like receptors and susceptibility to infectious disease. Lancet Infect Dis 2005;5:156-64. [Crossref] [PubMed]
- Martin SL, Desai S, Nanavati R, et al. Innate immune gene polymorphisms and their association with neonatal sepsis. Infect Genet Evol 2018;62:205-10. [Crossref] [PubMed]
- Kutukculer N, Yeniay BS, Aksu G, et al. Arg753Gln polymorphism of the human toll-like receptor-2 gene in children with recurrent febrile infections. Biochem Genet 2007;45:507-14. [Crossref] [PubMed]
- Lu H, Wen D, Wang X, et al. Host genetic variants in sepsis risk: a field synopsis and meta-analysis. Crit Care 2019;23:26. [Crossref] [PubMed]
- Abu-Maziad A, Schaa K, Bell EF, et al. Role of polymorphic variants as genetic modulators of infection in neonatal sepsis. Pediatr Res 2010;68:323-9. [Crossref] [PubMed]
- Sampath V, Mulrooney NP, Garland JS, et al. Toll-like receptor genetic variants are associated with Gram-negative infections in VLBW infants. J Perinatol 2013;33:772-7. [Crossref] [PubMed]
- Yuan FF, Marks K, Wong M, et al. Clinical relevance of TLR2, TLR4, CD14 and FcgammaRIIA gene polymorphisms in Streptococcus pneumoniae infection. Immunol Cell Biol 2008;86:268-70. [Crossref] [PubMed]
- Ahrens P, Kattner E, Köhler B, et al. Mutations of genes involved in the innate immune system as predictors of sepsis in very low birth weight infants. Pediatr Res 2004;55:652-6. [Crossref] [PubMed]
- Härtel C, Schultz C, Herting E, et al. Genetic association studies in VLBW infants exemplifying susceptibility to sepsis--recent findings and implications for future research. Acta Paediatr 2007;96:158-65. [Crossref] [PubMed]
- Świerzko AS, Szala-Poździej A, Kilpatrick DC, et al. Components of the lectin pathway of complement activation in paediatric patients of intensive care units. Immunobiology 2016;221:657-69. [Crossref] [PubMed]
- Fakhri D, Djauzi S, Murni TW, et al. Genetic polymorphism in postoperative sepsis after open heart surgery in infants. Asian Cardiovasc Thorac Ann 2016;24:326-31. [Crossref] [PubMed]
- Djuric O, Andjelkovic M, Vreca M, et al. Genetic variants in TNFA, LTA, TLR2 and TLR4 genes and risk of sepsis in patients with severe trauma: nested case-control study in a level-1 trauma centre in SERBIA. Injury 2021;52:419-25. [Crossref] [PubMed]
- Tang D, Kang R, Coyne CB, et al. PAMPs and DAMPs: signal 0s that spur autophagy and immunity. Immunol Rev 2012;249:158-75. [Crossref] [PubMed]
- Pashenkov MV, Murugina NE, Budikhina AS, et al. Synergistic interactions between NOD receptors and TLRs: Mechanisms and clinical implications. J Leukoc Biol 2019;105:669-80. [Crossref] [PubMed]
- Sampath V, Mulrooney N, Patel AL, et al. A Potential Role for the NOD1 Variant (rs6958571) in Gram-Positive Blood Stream Infection in ELBW Infants. Neonatology 2017;112:354-8. [Crossref] [PubMed]
- Tekin D, Dalgic N, Kayaalti Z, et al. Importance of NOD2/CARD15 gene variants for susceptibility to and outcome of sepsis in Turkish children. Pediatr Crit Care Med 2012;13:e73-7. [Crossref] [PubMed]
- Arcaroli J, Fessler MB, Abraham E. Genetic polymorphisms and sepsis. Shock 2005;24:300-12. [Crossref] [PubMed]
- Baldini M, Lohman IC, Halonen M, et al. A Polymorphism* in the 5' flanking region of the CD14 gene is associated with circulating soluble CD14 levels and with total serum immunoglobulin E. Am J Respir Cell Mol Biol 1999;20:976-83. [Crossref] [PubMed]
- Baier RJ, Loggins J, Yanamandra K. IL-10, IL-6 and CD14 polymorphisms and sepsis outcome in ventilated very low birth weight infants. BMC Med 2006;4:10. [Crossref] [PubMed]
- Esposito S, Zampiero A, Pugni L, et al. Genetic polymorphisms and sepsis in premature neonates. PLoS One 2014;9:e101248. [Crossref] [PubMed]
- Mustarim M, Yanwirasti Y, Jamsari J, et al. Association of Gene Polymorphism of Bactericidal Permeability Increasing Protein Rs4358188, Cluster of Differentiation 14 Rs2569190, Interleukin 1β Rs1143643 and Matrix Metalloproteinase-16 Rs2664349 with Neonatal Sepsis. Open Access Maced J Med Sci 2019;7:2728-33. [Crossref] [PubMed]
- Zhang AQ, Yue CL, Gu W, et al. Association between CD14 promoter -159C/T polymorphism and the risk of sepsis and mortality: a systematic review and meta-analysis. PLoS One 2013;8:e71237. [Crossref] [PubMed]
- Worthley DL, Bardy PG, Mullighan CG. Mannose-binding lectin: biology and clinical implications. Intern Med J 2005;35:548-55. [Crossref] [PubMed]
- Epstein J, Eichbaum Q, Sheriff S, et al. The collectins in innate immunity. Curr Opin Immunol 1996;8:29-35. [Crossref] [PubMed]
- De Pascale G, Cutuli SL, Pennisi MA, et al. The role of mannose-binding lectin in severe sepsis and septic shock. Mediators Inflamm 2013;2013:625803. [Crossref] [PubMed]
- Luo J, Xu F, Lu GJ, et al. Low mannose-binding lectin (MBL) levels and MBL genetic polymorphisms associated with the risk of neonatal sepsis: An updated meta-analysis. Early Hum Dev 2014;90:557-64. [Crossref] [PubMed]
- Özkan H, Köksal N, Çetinkaya M, et al. Serum mannose-binding lectin (MBL) gene polymorphism and low MBL levels are associated with neonatal sepsis and pneumonia. J Perinatol 2012;32:210-7. [Crossref] [PubMed]
- Koroglu OA, Onay H, Erdemir G, et al. Mannose-binding lectin gene polymorphism and early neonatal outcome in preterm infants. Neonatology 2010;98:305-12. [Crossref] [PubMed]
- Frakking FN, Brouwer N, van Eijkelenburg NK, et al. Low mannose-binding lectin (MBL) levels in neonates with pneumonia and sepsis. Clin Exp Immunol 2007;150:255-62. [Crossref] [PubMed]
- Dzwonek AB, Neth OW, Thiébaut R, et al. The role of mannose-binding lectin in susceptibility to infection in preterm neonates. Pediatr Res 2008;63:680-5. [Crossref] [PubMed]
- van der Zwet WC, Catsburg A, van Elburg RM, et al. Mannose-binding lectin (MBL) genotype in relation to risk of nosocomial infection in pre-term neonates in the neonatal intensive care unit. Clin Microbiol Infect 2008;14:130-5. [Crossref] [PubMed]
- Auriti C, Prencipe G, Inglese R, et al. Role of mannose-binding lectin in nosocomial sepsis in critically ill neonates. Hum Immunol 2010;71:1084-8. [Crossref] [PubMed]
- Badawy M, Mosallam DS, Saber D, et al. Use of Mannose-Binding Lectin Gene Polymorphisms and the Serum MBL Level for the Early Detection of Neonatal Sepsis. J Pediatr Genet 2018;7:150-7. [Crossref] [PubMed]
- Vassalli P. The pathophysiology of tumor necrosis factors. Annu Rev Immunol 1992;10:411-52. [Crossref] [PubMed]
- Wilson AG, Symons JA, McDowell TL, et al. Effects of a polymorphism in the human tumor necrosis factor alpha promoter on transcriptional activation. Proc Natl Acad Sci U S A 1997;94:3195-9. [Crossref] [PubMed]
- Sipahi T, Pocan H, Akar N. Effect of various genetic polymorphisms on the incidence and outcome of severe sepsis. Clin Appl Thromb Hemost 2006;12:47-54. [Crossref] [PubMed]
- Hedberg CL, Adcock K, Martin J, et al. Tumor necrosis factor alpha -- 308 polymorphism associated with increased sepsis mortality in ventilated very low birth weight infants. Pediatr Infect Dis J 2004;23:424-8. [Crossref] [PubMed]
- Härtel C, Hemmelmann C, Faust K, et al. Tumor necrosis factor-α promoter -308 G/A polymorphism and susceptibility to sepsis in very-low-birth-weight infants. Crit Care Med 2011;39:1190-5. [Crossref] [PubMed]
- Srinivasan L, Swarr DT, Sharma M, et al. Systematic Review and Meta-analysis: Gene Association Studies in Neonatal Sepsis. Am J Perinatol 2017;34:684-92. [PubMed]
- Allam G, Alsulaimani AA, Alzaharani AK, et al. Neonatal infections in Saudi Arabia: Association with cytokine gene polymorphisms. Cent Eur J Immunol 2015;40:68-77. [Crossref] [PubMed]
- Zhang M, Zhao Y, Liu Q. Tumor necrosis factor-α -308G/A and -238G/A polymorphisms are associated with increased risks of sepsis: evidence from an updated meta-analysis. APMIS 2017;125:459-67. [Crossref] [PubMed]
- Dinarello CA, Wolff SM. The role of interleukin-1 in disease. N Engl J Med 1993;328:106-13. [Crossref] [PubMed]
- Berner R, Niemeyer CM, Leititis JU, et al. Plasma levels and gene expression of granulocyte colony-stimulating factor, tumor necrosis factor-alpha, interleukin (IL)-1beta, IL-6, IL-8, and soluble intercellular adhesion molecule-1 in neonatal early onset sepsis. Pediatr Res 1998;44:469-77. [Crossref] [PubMed]
- Varljen T, Sekulovic G, Rakic O, et al. Genetic variant rs16944 in IL1B gene is a risk factor for early-onset sepsis susceptibility and outcome in preterm infants. Inflamm Res 2020;69:155-7. [Crossref] [PubMed]
- Zhang AQ, Pan W, Gao JW, et al. Associations between interleukin-1 gene polymorphisms and sepsis risk: a meta-analysis. BMC Med Genet 2014;15:8. [Crossref] [PubMed]
- Balding J, Healy CM, Livingstone WJ, et al. Genomic polymorphic profiles in an Irish population with meningococcaemia: is it possible to predict severity and outcome of disease? Genes Immun 2003;4:533-40. [Crossref] [PubMed]
- Treszl A, Kocsis I, Szathmári M, et al. Genetic variants of TNF-[FC12]a, IL-1beta, IL-4 receptor [FC12]a-chain, IL-6 and IL-10 genes are not risk factors for sepsis in low-birth-weight infants. Biol Neonate 2003;83:241-5. [Crossref] [PubMed]
- Fajgenbaum DC, June CH. Cytokine Storm. N Engl J Med 2020;383:2255-73. [Crossref] [PubMed]
- Kilpinen S, Hulkkonen J, Wang XY, et al. The promoter polymorphism of the interleukin-6 gene regulates interleukin-6 production in neonates but not in adults. Eur Cytokine Netw 2001;12:62-8. [PubMed]
- Reiman M, Kujari H, Ekholm E, et al. Interleukin-6 polymorphism is associated with chorioamnionitis and neonatal infections in preterm infants. J Pediatr 2008;153:19-24. [Crossref] [PubMed]
- Michalek J, Svetlikova P, Fedora M, et al. Interleukin-6 gene variants and the risk of sepsis development in children. Hum Immunol 2007;68:756-60. [Crossref] [PubMed]
- Harding D, Dhamrait S, Millar A, et al. Is interleukin-6 -174 genotype associated with the development of septicemia in preterm infants? Pediatrics 2003;112:800-3. [Crossref] [PubMed]
- Zidan HE, Elbehedy RM, Azab SF. IL6-174 G/C gene polymorphism and its relation to serum IL6 in Egyptian children with community-acquired pneumonia. Cytokine 2014;67:60-4. [Crossref] [PubMed]
- Göpel W, Härtel C, Ahrens P, et al. Interleukin-6-174-genotype, sepsis and cerebral injury in very low birth weight infants. Genes Immun 2006;7:65-8. [Crossref] [PubMed]
- Gao JW, Zhang AQ, Pan W, et al. Association between IL-6-174G/C polymorphism and the risk of sepsis and mortality: a systematic review and meta-analysis. PLoS One 2015;10:e0118843. [Crossref] [PubMed]
- Wacharasint P, Nakada TA, Boyd JH, et al. AA genotype of IL-8 -251A/T is associated with low PaO(2)/FiO(2) in critically ill patients and with increased IL-8 expression. Respirology 2012;17:1253-60. [Crossref] [PubMed]
- Hull J, Thomson A, Kwiatkowski D. Association of respiratory syncytial virus bronchiolitis with the interleukin 8 gene region in UK families. Thorax 2000;55:1023-7. [Crossref] [PubMed]
- Georgitsi MD, Vitoros V, Panou C, et al. Individualized significance of the -251 A/T single nucleotide polymorphism of interleukin-8 in severe infections. Eur J Clin Microbiol Infect Dis 2016;35:563-70. [Crossref] [PubMed]
- Amaya MP, Criado L, Blanco B, et al. Polymorphisms of pro-inflammatory cytokine genes and the risk for acute suppurative or chronic nonsuppurative apical periodontitis in a Colombian population. Int Endod J 2013;46:71-8. [Crossref] [PubMed]
- Hu D, Wang H, Huang X, et al. Investigation of association between IL-8 serum levels and IL8 polymorphisms in Chinese patients with sepsis. Gene 2016;594:165-70. [Crossref] [PubMed]
- Fu P, Xie S, Zhang X. IL-8 gene locus is associated with risk, severity and 28-day mortality of sepsis in a Chinese population. Clin Exp Med 2019;19:571-6. [Crossref] [PubMed]
- Schaaf BM, Boehmke F, Esnaashari H, et al. Pneumococcal septic shock is associated with the interleukin-10-1082 gene promoter polymorphism. Am J Respir Crit Care Med 2003;168:476-80. [Crossref] [PubMed]
- Emonts M, Vermont CL, Houwing-Duistermaat JJ, et al. Polymorphisms in PARP, IL1B, IL4, IL10, C1INH, DEFB1, and DEFA4 in meningococcal disease in three populations. Shock 2010;34:17-22. [Crossref] [PubMed]
- Pan W, Zhang AQ, Yue CL, et al. Association between interleukin-10 polymorphisms and sepsis: a meta-analysis. Epidemiol Infect 2015;143:366-75. [Crossref] [PubMed]
- Levy O, Martin S, Eichenwald E, et al. Impaired innate immunity in the newborn: newborn neutrophils are deficient in bactericidal/permeability-increasing protein. Pediatrics 1999;104:1327-33. [Crossref] [PubMed]
- Levin M, Quint PA, Goldstein B, et al. Recombinant bactericidal/permeability-increasing protein (rBPI21) as adjunctive treatment for children with severe meningococcal sepsis: a randomised trial. rBPI21 Meningococcal Sepsis Study Group. Lancet 2000;356:961-7. [Crossref] [PubMed]
- Michalek J, Svetlikova P, Fedora M, et al. Bactericidal permeability increasing protein gene variants in children with sepsis. Intensive Care Med 2007;33:2158-64. [Crossref] [PubMed]
- Khokha R, Murthy A, Weiss A. Metalloproteinases and their natural inhibitors in inflammation and immunity. Nat Rev Immunol 2013;13:649-65. [Crossref] [PubMed]
- Elkington PT, O'Kane CM, Friedland JS. The paradox of matrix metalloproteinases in infectious disease. Clin Exp Immunol 2005;142:12-20. [Crossref] [PubMed]
- Hadchouel A, Decobert F, Franco-Montoya ML, et al. Matrix metalloproteinase gene polymorphisms and bronchopulmonary dysplasia: identification of MMP16 as a new player in lung development. PLoS One 2008;3:e3188. [Crossref] [PubMed]
- Watanabe K, Lam G, Keresztes RS, et al. Lipopolysaccharides decrease angiotensin converting enzyme activity expressed by cultured human endothelial cells. J Cell Physiol 1992;150:433-9. [Crossref] [PubMed]
- Chawla LS, Chen S, Bellomo R, et al. Angiotensin converting enzyme defects in shock: implications for future therapy. Crit Care 2018;22:274. [Crossref] [PubMed]
- Rigat B, Hubert C, Alhenc-Gelas F, et al. An insertion/deletion polymorphism in the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest 1990;86:1343-6. [Crossref] [PubMed]
- Dou XM, Cheng HJ, Meng L, et al. Correlations between ACE single nucleotide polymorphisms and prognosis of patients with septic shock. Biosci Rep 2017;37:BSR20170145. [Crossref] [PubMed]
- Hou X, Zhang P, Nie W, et al. Association between angiotensin-converting enzyme I/D polymorphism and sepsis: A meta-analysis. J Renin Angiotensin Aldosterone Syst 2015;16:415-21. [Crossref] [PubMed]
- Spiegler J, Gilhaus A, Konig IR, et al. Polymorphisms in the Renin-Angiotensin system and outcome of very-low-birthweight infants. Neonatology 2010;97:10-4. [Crossref] [PubMed]
- John Baier R, Loggins J, Yanamandra K. Angiotensin converting enzyme insertion/deletion polymorphism does not alter sepsis outcome in ventilated very low birth weight infants. J Perinatol 2005;25:205-9. [Crossref] [PubMed]
- Cogulu O, Onay H, Uzunkaya D, et al. Role of angiotensin-converting enzyme gene polymorphisms in children with sepsis and septic shock. Pediatr Int 2008;50:477-80. [Crossref] [PubMed]
- Jarahzadeh MH, Jafari M, Seifi-Shalamzari N, et al. Association of PAI-1 4G/5G and ACE I/D Polymorphisms with Susceptibility to Pediatric Sepsis: Evidence from a Meta-Analysis. Fetal Pediatr Pathol 2022;41:242-58. [Crossref] [PubMed]
- Salgado DR, Rocco JR, Silva E, et al. Modulation of the renin-angiotensin-aldosterone system in sepsis: a new therapeutic approach? Expert Opin Ther Targets 2010;14:11-20. [Crossref] [PubMed]
- Lee HW, Suh JK, Jang E, et al. Effect of angiotensin converting enzyme inhibitor and angiotensin II receptor blocker on the patients with sepsis. Korean J Intern Med 2021;36:371-81. [Crossref] [PubMed]
- Kostakoglu U, Topcu A, Atak M, et al. The protective effects of angiotensin-converting enzyme inhibitor against cecal ligation and puncture-induced sepsis via oxidative stress and inflammation. Life Sci 2020;241:117051. [Crossref] [PubMed]
- Bitker L, Burrell LM. Classic and Nonclassic Renin-Angiotensin Systems in the Critically Ill. Crit Care Clin 2019;35:213-27. [Crossref] [PubMed]
- Esmon CT. Inflammation and thrombosis. J Thromb Haemost 2003;1:1343-8. [Crossref] [PubMed]
- Lao TT, Yin JA, Yuen PM. Coagulation and anticoagulation systems in newborns--correlation with their mothers at delivery. Lower levels of anticoagulants and fibrinolytic activity in the newborn. Gynecol Obstet Invest 1990;29:181-4. [Crossref] [PubMed]
- Carcao MD, Blanchette VS, Dean JA, et al. The Platelet Function Analyzer (PFA-100): a novel in-vitro system for evaluation of primary haemostasis in children. Br J Haematol 1998;101:70-3. [Crossref] [PubMed]
- Härtel C, König I, Köster S, et al. Genetic polymorphisms of hemostasis genes and primary outcome of very low birth weight infants. Pediatrics 2006;118:683-9. [Crossref] [PubMed]
- Ariëns RA, Lai TS, Weisel JW, et al. Role of factor XIII in fibrin clot formation and effects of genetic polymorphisms. Blood 2002;100:743-54. [Crossref] [PubMed]
- Ariëns RA, Philippou H, Nagaswami C, et al. The factor XIII V34L polymorphism accelerates thrombin activation of factor XIII and affects cross-linked fibrin structure. Blood 2000;96:988-95. [Crossref] [PubMed]
- Texereau J, Pene F, Chiche JD, et al. Importance of hemostatic gene polymorphisms for susceptibility to and outcome of severe sepsis. Crit Care Med 2004;32:S313-9. [Crossref] [PubMed]
- Shi Q, Mu X, Hong L, et al. SERPINE1 rs1799768 polymorphism contributes to sepsis risk and mortality. J Renin Angiotensin Aldosterone Syst 2015;16:1218-24. [Crossref] [PubMed]
- Shi C, Sui Z, Li L, et al. No Association of SERPINE1 -675 Polymorphism With Sepsis Susceptibility: A Meta-Analysis. Medicine (Baltimore) 2015;94:e1173. [Crossref] [PubMed]
- Li L, Nie W, Zhou H, et al. Association between plasminogen activator inhibitor-1 -675 4G/5G polymorphism and sepsis: a meta-analysis. PLoS One 2013;8:e54883. [Crossref] [PubMed]
- Esmon CT. The protein C pathway. Chest 2003;124:26S-32S. [Crossref] [PubMed]
- Taylor FB Jr, Stearns-Kurosawa DJ, Kurosawa S, et al. The endothelial cell protein C receptor aids in host defense against Escherichia coli sepsis. Blood 2000;95:1680-6. [Crossref] [PubMed]
- Liang Y, Huang X, Jiang Y, et al. Endothelial protein C receptor polymorphisms and risk of sepsis in a Chinese population. J Int Med Res 2017;45:504-13. [Crossref] [PubMed]
- Vassiliou AG, Maniatis NA, Kotanidou A, et al. Endothelial protein C receptor polymorphisms and risk of severe sepsis in critically ill patients. Intensive Care Med 2013;39:1752-9. [Crossref] [PubMed]
- Opal SM, Dellinger RP, Vincent JL, et al. The next generation of sepsis clinical trial designs: what is next after the demise of recombinant human activated protein C?*. Crit Care Med 2014;42:1714-21. [Crossref] [PubMed]
- Fiusa MM, Carvalho-Filho MA, Annichino-Bizzacchi JM, et al. Causes and consequences of coagulation activation in sepsis: an evolutionary medicine perspective. BMC Med 2015;13:105. [Crossref] [PubMed]
- Bikle DD. Vitamin D metabolism, mechanism of action, and clinical applications. Chem Biol 2014;21:319-29. [Crossref] [PubMed]
- Dusso AS, Brown AJ, Slatopolsky E, Vitamin D. Am J Physiol Renal Physiol 2005;289:F8-28. [Crossref] [PubMed]
- Rukin NJ, Strange RC. What are the frequency, distribution, and functional effects of vitamin D receptor polymorphisms as related to cancer risk? Nutr Rev 2007;65:S96-101. [Crossref] [PubMed]
- Das B, Patra S, Behera C, et al. Genotyping of vitamin D receptor gene polymorphisms using mismatched amplification mutation assay in neonatal sepsis patients of Odisha, eastern India. Infect Genet Evol 2016;45:40-7. [Crossref] [PubMed]
- Zeljic K, Elkilany A, Supic G, et al. Vitamin D receptor gene polymorphisms association with the risk of sepsis and mortality. Int J Immunogenet 2017;44:129-34. [Crossref] [PubMed]
- Tayel SI, Soliman SE, Elsayed HM. Vitamin D deficiency and vitamin D receptor variants in mothers and their neonates are risk factors for neonatal sepsis. Steroids 2018;134:37-42. [Crossref] [PubMed]
- He D, Lu X, Li W, et al. Vitamin D Receptor Is a Sepsis-Susceptibility Gene in Chinese Children. Med Sci Monit 2021;27:e932518. [Crossref] [PubMed]
- Xiao L, Que S, Mu L, et al. The relationship between vitamin D receptor gene and TREM-1 gene polymorphisms and the susceptibility and prognosis of neonatal sepsis. J Clin Lab Anal 2022;36:e24405. [Crossref] [PubMed]
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell 2007;128:635-8. [Crossref] [PubMed]
- Berger SL, Kouzarides T, Shiekhattar R, et al. An operational definition of epigenetics. Genes Dev 2009;23:781-3. [Crossref] [PubMed]
- Cross D, Drury R, Hill J, et al. Epigenetics in Sepsis: Understanding Its Role in Endothelial Dysfunction, Immunosuppression, and Potential Therapeutics. Front Immunol 2019;10:1363. [Crossref] [PubMed]
- Beltrán-García J, Osca-Verdegal R, Romá-Mateo C, et al. Epigenetic biomarkers for human sepsis and septic shock: insights from immunosuppression. Epigenomics 2020;12:617-46. [Crossref] [PubMed]
- Weiterer S, Uhle F, Lichtenstern C, et al. Sepsis induces specific changes in histone modification patterns in human monocytes. PLoS One 2015;10:e0121748. [Crossref] [PubMed]
- Bermick J, Gallagher K, denDekker A, et al. Chorioamnionitis exposure remodels the unique histone modification landscape of neonatal monocytes and alters the expression of immune pathway genes. FEBS J 2019;286:82-109. [Crossref] [PubMed]
- Wang Q, Wang C, Li N, et al. Condensin Smc4 promotes inflammatory innate immune response by epigenetically enhancing NEMO transcription. J Autoimmun 2018;92:67-76. [Crossref] [PubMed]
- Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet 2008;9:465-76. [Crossref] [PubMed]
- Binnie A, Walsh CJ, Hu P, et al. Epigenetic Profiling in Severe Sepsis: A Pilot Study of DNA Methylation Profiles in Critical Illness. Crit Care Med 2020;48:142-50. [Crossref] [PubMed]
- Lorente-Sorolla C, Garcia-Gomez A, Català-Moll F, et al. Inflammatory cytokines and organ dysfunction associate with the aberrant DNA methylome of monocytes in sepsis. Genome Med 2019;11:66. [Crossref] [PubMed]
- Lorente-Pozo S, Navarrete P, Garzón MJ, et al. DNA Methylation Analysis to Unravel Altered Genetic Pathways Underlying Early Onset and Late Onset Neonatal Sepsis. A Pilot Study. Front Immunol 2021;12:622599. [Crossref] [PubMed]
- Dhas DB, Ashmi AH, Bhat BV, et al. Comparison of genomic DNA methylation pattern among septic and non-septic newborns - An epigenome wide association study. Genom Data 2015;3:36-40. [Crossref] [PubMed]
- Tendl KA, Schulz SM, Mechtler TP, et al. DNA methylation pattern of CALCA in preterm neonates with bacterial sepsis as a putative epigenetic biomarker. Epigenetics 2013;8:1261-7. [Crossref] [PubMed]
- Li LC. Chromatin remodeling by the small RNA machinery in mammalian cells. Epigenetics 2014;9:45-52. [Crossref] [PubMed]
- Cheng L, Nan C, Kang L, et al. Whole blood transcriptomic investigation identifies long non-coding RNAs as regulators in sepsis. J Transl Med 2020;18:217. [Crossref] [PubMed]
- Scicluna BP, Uhel F, van Vught LA, et al. The leukocyte non-coding RNA landscape in critically ill patients with sepsis. Elife 2020;9:e58597. [Crossref] [PubMed]
- Pellegrina DVDS, Severino P, Barbeiro HV, et al. Insights into the Function of Long Noncoding RNAs in Sepsis Revealed by Gene Co-Expression Network Analysis. Noncoding RNA 2017;3:5. [Crossref] [PubMed]
- Wang X, Wang X, Liu X, et al. miR-15a/16 are upreuglated in the serum of neonatal sepsis patients and inhibit the LPS-induced inflammatory pathway. Int J Clin Exp Med 2015;8:5683-90. [PubMed]
- Chen J, Jiang S, Cao Y, et al. Altered miRNAs expression profiles and modulation of immune response genes and proteins during neonatal sepsis. J Clin Immunol 2014;34:340-8. [Crossref] [PubMed]
- Gluckman PD, Hanson MA, Cooper C, et al. Effect of in utero and early-life conditions on adult health and disease. N Engl J Med 2008;359:61-73. [Crossref] [PubMed]
- Khera AV, Chaffin M, Aragam KG, et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 2018;50:1219-24. [Crossref] [PubMed]
- Mavaddat N, Michailidou K, Dennis J, et al. Polygenic Risk Scores for Prediction of Breast Cancer and Breast Cancer Subtypes. Am J Hum Genet 2019;104:21-34. [Crossref] [PubMed]
- Lewis CM, Vassos E. Polygenic risk scores: from research tools to clinical instruments. Genome Med 2020;12:44. [Crossref] [PubMed]
- Lu H, Wen D, Sun J, et al. Polygenic Risk Score for Early Prediction of Sepsis Risk in the Polytrauma Screening Cohort. Front Genet 2020;11:545564. [Crossref] [PubMed]
- Engoren M, Jewell ES, Douville N, et al. Genetic variants associated with sepsis. PLoS One 2022;17:e0265052. [Crossref] [PubMed]
- D'Urso S, Rajbhandari D, Peach E, et al. Septic Shock: A Genomewide Association Study and Polygenic Risk Score Analysis. Twin Res Hum Genet 2020;23:204-13. [Crossref] [PubMed]
- Lee J, Son W, Hong J, et al. Down-regulation of TNF-α via macrophage-targeted RNAi system for the treatment of acute inflammatory sepsis. J Control Release 2021;336:344-53. [Crossref] [PubMed]
- Liu AC, Patel K, Vunikili RD, et al. Sepsis in the era of data-driven medicine: personalizing risks, diagnoses, treatments and prognoses. Brief Bioinform 2020;21:1182-95. [Crossref] [PubMed]
- Lin PI, Vance JM, Pericak-Vance MA, et al. No gene is an island: the flip-flop phenomenon. Am J Hum Genet 2007;80:531-8. [Crossref] [PubMed]
- Watson JD. Celebrating the genetic jubilee: a conversation with James D. Watson. Interviewed by John Rennie. Sci Am 2003;288:66-9. [Crossref] [PubMed]


