
Modeling congenital brain malformations with brain organoids: a narrative review
Introduction
The human brain is one of the most complex organs in the body, and its development occurs through a series of processes, including neurogenesis, cell migration, neuronal maturation, and neural circuit formation (1). The nervous system originates from the ectoderm, with the neural tube and neural crest giving rise to the central and peripheral nervous systems, respectively. The neocortex development starts with the expansion of neuroepithelial cells at the ventricular zone (VZ), which gives rise to radial glial (RG) cells. RG cells have the capacity for self-renewal to expand the neural progenitor cells (NPCs) pool and can differentiate into intermediate progenitor cells or neurons directly (2,3). The newborn neurons then migrate to their final destinations in the different layers of the cerebral cortex (4,5). Dysfunctions of early neurodevelopment, including neurogenesis and neural migration, have been associated with structural malformations of the human brain.
Congenital brain malformations comprise a diverse group of neurodevelopmental disorders that occur during early embryonic development and are caused by genetic or environmental factors (6). The pathogenesis of congenital brain malformations remains largely unclear due to the lack of disease models based on the human genetic background. Most studies choose rodent animals as the disease model to explore the pathophysiology of human brain malformations. However, rodent animal brains are lissencephaly (LIS) and short of outer radial glial cells (oRGs), posing a formidable barrier to recapitulate human brain diseases (7). Moreover, single-cell transcriptomics showed extensive differences in cell types and gene expression between humans and rodents (8). This emphasizes the importance of species-specific models.
Human pluripotent stem cells (hPSCs), including human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs), opened a new avenue to model human diseases in vitro (9). hPSCs-derived 3-dimensional (3D) aggregates named brain organoids have similarities with human brains in cell types, cytoarchitectures, and spatial architectures. Therefore, brain organoids have become potential tools for exploring the mechanism of congenital human brain malformations (10).
Here, we summarize the development of methodologies to differentiate human brain organoids and their applications in brain disease modeling, especially for congenital brain malformations, such as microcephaly, macrocephaly, LIS, and periventricular nodular heterotopia (PH). Furthermore, we also discuss future perspectives for advancing current brain organoid technologies to expand their applications. We present the following article in accordance with the Narrative Review reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-22-239/rc).
Methods
We performed a comprehensive search using PubMed and Web of Science’s Core Collection for literature published from July 1, 2000 to July 1, 2022. Keywords included terms related to the technology (brain organoid/cerebral organoid) and the disease (congenital brain malformations/structural brain abnormalities), as well as names of individual malformations (microcephaly/macrocephaly/LIS/PH). The detailed search strategy is summarized in Table 1. The literature research was supplemented by reviewing references within individual papers.
Table 1
| Items | Specification |
|---|---|
| Date of search | The first search was conducted on November 1, 2021. The second search was conducted on July 1, 2022 |
| Databases and other sources searched | PubMed and Web of Science’s core collection |
| Search terms used | Combinations of terms related to the technology (brain organoid/cerebral organoid) and the disease (congenital brain malformation/structural brain abnormalities), as well as names of individual malformations (microcephaly/macrocephaly/lissencephaly/periventricular nodular heterotopia) appearing in article title, abstract or medical subject headings |
| Timeframe | Published between July 2000 to July 2022 |
| Inclusion and exclusion criteria | All study types published in the English language were included |
| Selection process | The search was conducted by two authors (XSJ and XLJ) independently. Any disagreement between the two authors was resolved by a third author (MX) |
Brain organoids methodologies
In general, there are two approaches to generating brain organoids: unguided and guided methods (Figure 1A). The protocol of generating unguided brain organoids, or cerebral organoids, has continued to advance in the last decade. The first embryonic stem cell (ESC)-derived cortical neuroepithelia, which was considered a 2.5-dimensional (2.5 D) brain model, was established in 2008 using a technique known as SFEBq [serum-free floating culture of embryoid body (EB)-like aggregates] (11). The self-organized tissue showed apical-basal polarization and mimicked structural aspects of the human cerebral cortex. Then, Eiraku et al. (12) recapitulated optic-cup development with dissolved Matrigel, which was the first instance of neural tissue being fully self-organized in vitro. In 2013, Lancaster et al. (13) developed the first 3D culture system for cerebral organoids. Neuroectoderm was generated from EBs and embedded in droplets of Matrigel, which could provide a scaffold for complex tissue growth. Then, the droplets were transferred to a spinning bioreactor or orbital shaker to continue the culture, and human brain organoids formed within two months (Figure 1B). This method generated unguided brain organoids, or cerebral organoids, which exhibited a variety of cell lineage identities, including forebrain, midbrain, and hindbrain.
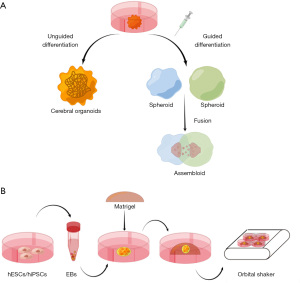
Although the cell-type diversity in cerebral organoids sheds light on the interactions of different brain regions, the differentiation processes are fully dependent on cell spontaneous and intrinsic signals, leading to high variability and heterogeneity in different cell lines. Thus, small molecules were added during the differentiation process to form certain brain regions. For example, SB-431542, IWR1, and Dorsomorphin were added to induce forebrain organoids formation (14). WNT3A, SHH, and Purmorphamine were used to guide thalamic organoids (15). Recently, distinct region-specific organoids were fused to form an “assembloid” to study interregional interactions. For instance, Xiang et al. (16) studied interneuron migration by fusing medial ganglionic eminence organoids with cortical organoids. Furthermore, vascularized human brain organoids were generated by co-culturing them with endothelial cells or introducing hESCs expressing the endothelial-inducing transcription factor EVT2 (17,18). These advances provided precision tools to model brain development and more complex neurological disorders.
Modeling congenital brain malformations using organoids
So far, brain organoids have been used to study various kinds of congenital brain malformations. We mainly discuss the pioneering studies in microcephaly, macrocephaly, LIS, and PH (Table 2; Figure 2).
Table 2
| Brain malformation | Gene | PSC | Brain region modeled | Phenotype of organoids | Molecular mechanism | Ref. |
|---|---|---|---|---|---|---|
| Microcephaly | CDK5RAP2 | hiPSC | Whole brain | Smaller neural tissues, premature neural differentiation | – | (13) |
| Microcephaly | CPAP | hiPSC | Whole brain | Reduced neural epithelial tissues, increased numbers, and longer cilia | Loss of CC5 domain | (19) |
| Microcephaly | ASPM | hiPSC | Cerebral cortex | Less organized neuroepithelium | – | (20) |
| Microcephaly | WDR62 | hiPSC or hESC | Whole brain | Retarded cilium disassembly, long cilium, and delayed cell cycle progression | Disruption of WDR62-CEP170-KIF2A pathway | (21) |
| Microcephaly | KNL1 | hESC | Whole brain | Reduced proliferation of NPCs and premature differentiation | Generation of an exonic splicing silencer site | (22) |
| Microcephaly | IER3IP1 | hESC | Whole brain | Neural progenitor loss | – | (23) |
| Microcephaly | PTEN | hPSCs | Whole brain | Reduced NPCs proliferation, promoted neuronal differentiation | Reduction of AKT pathway activity | (24) |
| Microcephaly | 16p11.2 DUP | hiPSC | Cerebral cortex | Smaller in size | – | (25) |
| Microcephaly | AUTS2 | hiPSC | Whole brain | Reduced NPCs proliferation, disrupted NPCs polarity | Alteration in WNT-β-Catenin signaling | (26) |
| Macrocephaly | PTEN | hiPSC | Telencephalon | Increased organoid size; decreased cell cycle length; overproduction of GABAergic neurons | – | (27) |
| Macrocephaly | PTEN | hESC | Whole brain | Expanded VZ/SVZ, increased size | Increased AKT signaling | (28) |
| Macrocephaly | RAN39b | hiPSC, hESC | Whole brain | Enlarged organoid sizes, increased NPC proliferation, a transient delay of differentiation, and increased output of neuronal production | Upregulation of PI3K-AKT-mTOR signaling activity | (7) |
| Macrocephaly | CHD8 | hESC | Whole brain | Enlarged organoid sizes, increased NPC proliferation, imbalanced excitatory and inhibitory neuronal production | Affected mRNA metabolism | (29) |
| Macrocephaly | STRADA | hiPSC | Cerebral cortex | Enlarged size, delayed neurogenesis, increased outer radial glia | mTOR pathway hyperactivation | (30) |
| Lissencephaly | 17p13.3 DEL | hiPSC | Whole brain | Decreased vertical divisions, reduced migration speed, and overproduction of deep-layer neurons | – | (31) |
| Lissencephaly | 17p13.3 DEL | hiPSC | Forebrain | Reduced size and expansion rate | Disruption of N-cadherin/β-catenin/Wnt signaling | (32) |
| Microlissencep-haly | KATNB1 | hiPSC | Whole brain | TuJ1+ neurons fail to migrate out of the proliferating fields | Loss of p80 causes improper MT organization | (33) |
| Periventricular nodular heterotopia | OLEKHG6 | hiPSC | Whole brain | Impaired ventricular surface and induced heterotopic neurons clustering | Activation of the small GTPase RhoA | (34) |
| Periventricular nodular heterotopia | DCHS1, FAT4 | hiPSC | Whole brain | Defective NPC morphology and neuronal migration dynamics | – | (35) |
| Periventricular nodular heterotopia | ECE2 | hiPSC | Whole brain | Ectopic localization of neural progenitors and neurons | Changes in actin and microtubule cytoskeleton | (2) |
| Periventricular nodular heterotopia | GNG5 | hiPSC | Whole brain | Premature delamination and neuronal migration defects | – | (36) |
PSC, pluripotent stem cell; hiPSCs, human induced pluripotent stem cells; hESCs, human embryonic stem cells; NPC, neuronal progenitor cells; CP, cortical plate; VZ, ventricular zone; SVZ, subventricular zone; 3D, 3-dimensional.

Microcephaly
Microcephaly is defined as a head circumference of 3 standard deviations (SDs) or more below the mean for the gestational age and gender (37). Primary microcephaly is mostly caused by autosomal recessive mutations. So far, 27 genes have been reported to be involved in microcephaly. Centriole and spindle biogenesis and impaired DNA damage represent the two most common mechanisms that are negatively affected (38). Furthermore, perinatal viral infections account for up to 50% of cases of primary microcephaly (39), including Zika virus (ZIKV), herpes simplex virus 1 (HSV-1), and human cytomegalovirus (HCMV) (40).
Brain organoids have helped reveal the mechanisms of microcephaly and illuminated the surprising roles of centrosomes and primary cilia in regulating neurogenesis. Brain organoids derived from microcephaly patients with mutations in centrosome-associated genes such as CDK5RAP2 showed premature neuronal differentiation and a smaller size (13,22,24,26). Gabriel et al. (19) developed iPS-derived organoids which harbored a splice-site mutation in CPAP and observed longer cilia, retarded cilium disassembly, and delayed cell cycle re-entry leading to premature differentiation of NPCs. They suggested that cilia disassembly, cell cycle progression, and neuronal differentiation were causally related to each other, and cilium played a crucial role in NPCs maintenance. Li et al. (20) modeled the cellular defects of ASPM-dependent microcephaly by 3D brain organoids. They showed fewer oRGs which could not be recapitulated with rodents due to limited oRGs in their brains.
In addition to genetic conditions, brain organoids have been used to model microcephaly caused by neurotrophic pathogens. For example, ZIKV experimental exposure in brain organoids induced increased apoptosis and decreased proliferation of NPCs, leading to reduced organoid size (28,41,42). Recently, the upregulation of type I interferons was observed in ZIKV and HSV-1-infected organoids, which was not seen in 2D cultures, highlighting the superiority of brain organoids in disease modeling (43). Based on virus-infected brain organoids, several studies have tested compounds and drugs such as hippeastrine hydrobromide and duramycin for treating infected brain organoids, which rescued structural defects (44-48).
Macrocephaly
Macrocephaly is characterized by a head circumference of 2 SDs or more above the mean for the gestational age and gender (49). It can be used interchangeably with megalencephaly, a distinct term suggesting increased growth of the cerebral structure without hydrocephaly (50).
Increased proliferation of NPCs contributes to the expansion of the human cerebral cortex (51,52). Deletion of PTEN generated larger cerebral organoids, which displayed an expanded NPC pool and a transient neuronal differentiation delay in the fourth week (28). Similarly, Dang et al. (30) developed STRADA-mutant human cortical organoids, and they found enlarged sizes and increased neuroepithelial budding of these disease-specific organoids after 2 weeks. These organoids also exhibited increased cell proliferation, early apoptosis, and increased extraluminal primary cilia. They also displayed increased oRGs in the twelfth week, suggesting another potential mechanism for macrocephaly.
Interestingly, a consistent positive correlation between macrocephaly and the severity of autism spectrum disorder (ASD) has been found using 3D neural cultures. Brain organoids derived from ASD individuals showed features of altered neurodevelopment, including decreased cell cycle length, increased cell proliferation, and unbalanced inhibitory neuron differentiation (27). Villa et al. (29) found that CHD8 mutant organoids induced from patients with macrocephaly showed a cell-autonomous sustained proliferation of neural precursors. The overgrowth of neural precursors disrupted the balance between excitatory and inhibitory neuronal production, with delayed production of excitatory neurons. In terms of the mechanism, RAB39b-mutant organoids revealed the activation of PI3K-AKT-mTOR signaling, which contributed to macrocephaly and autistic-like behaviors (7). Altogether, brain organoids represented a novel method to investigate the molecular and anatomical paradigms of macrocephaly. The findings were consistent with the notion that individuals with macrocephaly had poor ASD clinical outcomes.
LIS
LIS, also called smooth brain, is a disorder involved in neuronal proliferation and migration characterized by a reduced number or absence of gyri and sulci (53). The identified genetic causes of LIS include mutations in PAFAH1B1, YWHAE, DCX, TUBA1A, and RELN, most of which encode cytoskeletal proteins (54). Miller-Dieker syndrome (MDS), caused by a heterozygous deletion of 17p13.3 involving PAFAH1B1 and YWHAE, is the most severe form of LIS.
As the mouse brain is naturally lissencephalic and short of a separate outer subventricular zone (oSVZ), the phenotypes in Pafah1b1+/- mice are substantially milder than in human patients. For the investigation of cell-type-specific defects in LIS, cerebral organoids were cultured from MDS patient-derived iPSCs (31). The MDS organoids recapitulated several MDS phenotypes, such as a significantly smaller size of the brain. An early defect in the expansion of the neuroepithelial cells was observed due to increased apoptosis and decreased vertical divisions, which would reduce NPCs pools. Also, the MDS neurons showed reduced track speed and straightness during migration, which could be functionally rescued by compensatory duplication of wild-type chromosome 17 (31). In another study, disruption of the N-cadherin/β-catenin/Wnt signaling axis was shown in MDS-derived organoids, leading to an imbalance of proliferation and differentiation in cortical progenitors (32). The phenotype could be ameliorated by external activation of Wnt signaling.
Although brain organoids have been successfully used to recognize the pathogenesis in human LIS, real cortical folding remains unachieved so far in cortical organoids (9). There have been efforts to engineer neuroepithelial “wrinkling” or “pseudo-folding” during early differentiation, either by inducing enhanced proliferation of NPCs through genetic manipulation or by using internal mechanical constraint in a microfluidic device (28,55-57). However, they do not lead to the formation of gyrification at the human brain level (58). According to statistics, the degree of folding scales with the surface area and the thickness of the cortical plate (CP), while organoids may be too small to achieve this (59,60). Better approaches may be needed to engineer organoids recapitulating the folding of the human neocortex.
PH
PH is another common neuronal migration disorder. It is characterized by a subset of cortical neurons that fail to locate correctly within the cerebral cortex and instead lining their sites of production as nodules (2). A few genes have been shown to cause PH, and high-throughput analysis has demonstrated extreme genetic heterogeneity (61).
O’Neill et al. (34) generated cerebral organoids from iPSCs derived from patients with a mutation in PLEKHG6. Ectopic clusters composed of neural progenitors and neurons were identified at the ventricular surfaces. The mutation also impaired the neuroepithelial lining and adherent junction belt. And functional analysis showed that the dysfunction of oRGs played an important role in the pathogenesis of PH. Similarly, mutations in DCHS1, FAT4, ENE2, and GNG5 caused disrupted progenitor morphology and resulted in defective neuronal migration dynamics in brain organoids (2,35,36). Despite the altered neuronal migratory trajectories that have been demonstrated, an understanding of the molecular role of these genes during cortical development is only beginning to emerge.
Advances, limitations, and future perspectives of brain organoids
Brain organoids have proven to be a powerful system to understand congenital brain abnormalities because they provide a direct read-out for brain malformations and bridge the gap between 2D cultures and human brains. First and foremost, originating from human patients has made them better reflect the genetic background and physiology of the human brain than animal models. Also, compared with 2D cell cultures, brain organoids generate a complex architecture containing different neuronal subtypes, with proper spatial alignment and cell-cell interactions, which better recapitulates the dynamic developmental process of human brains.
Although brain organoids have greatly expanded our ability to model congenital brain malformations, there are still some hurdles to overcome (Table 3). First, fully organized six-layered laminar neocortical architecture and cortical folding remain unrealized, and one of the reasons is the insufficient oxygen and nutrient diffusion to the inner part of organoids. Many methodologies, such as modifications of EB size, combination with bioengineering constructs, and air-liquid interface culture, have been built to maintain oxygen and nutrient supply (57,62,63). Organoids with larger continuous cortical lobes were generated using a microscale internal scaffold to shape the organoids at the EB stage (64). Furthermore, the engineering of vascularized human brain organoids could supply oxygen and nutrient to the central region of organoids (17). Transplanting vascularized brain organoids into the mouse brain has been shown to generate functional neuronal networks and blood vessels (65).
Table 3
| Limitation | Current solution | Future perspective |
|---|---|---|
| Insufficient oxygen and nutrient diffusion | Modifications of EB size, combined with bioengineering constructs and air-liquid interface culture; vessel organoids; transplantation of organoids into animals | Vascularized organoids; organoids with six-layered laminar neocortical architecture and cortical folding |
| Incomplete cellular diversity | Transplant pre-differentiated cells to brain organoids; timed exposure to specific growth factors and hormones | Mature glia subpopulations in the same brain organoid |
| Low reproducibility | Use of micro-scaffolds, the addition of exogenous patterning factors, and the use of mini-spinning bioreactors; minimized use of variable ingredients; quantitative characterization | Increased reproducibility |
EB, embryoid body.
Second, although some groups have developed astrocytes and microglia from iPSCs and transplanted them to brain organoids (66,67), morphologically and functionally mature glia subpopulations developing all in the same brain organoid are still out of reach. Recently, oligodendrocytes and myelin were generated in cortical spheroids and were used to recapitulate the pathologic process of a genetic myelin disorder (68). Future studies need to focus on complex structures underlying functional neuron-glia interactions.
Lastly, as the organoid differentiation protocols, especially the unguided methods, rely on spontaneous morphogenesis and intrinsic differentiation capacities of hPSCs, they suffer from batch-to-batch heterogeneity. Research has shown that inconsistent neural induction efficiency could be a main source of variability (69). Attempts to increase the homogeneity included the use of micro-scaffolds to arrange cells in an organ-like configuration (64), the addition of exogenous patterning factors to generate region-specific organoids (70,71), and the use of mini-spinning bioreactors with minimized volumes of variable ingredients and better-controlled conditions (72). In addition, newly generated organoids need to be qualitatively characterized to show consistent results in multiple batches (73).
Moving forward, integrating brain organoids with state-of-the-art technologies, such as lineage-coupled single-cell transcriptomics, long-term live imaging, and automated read-outs for high-throughput analyses, will help to exploit organoids to their full potential in clinical settings and translate well to patients’ bedsides. The molecular diagnosis and treatment of congenital brain malformations are challenging for physicians, and most genes have not been characterized in depth. By combining CRISPR-Cas9 with lineage barcode, Esk et al. (23) established a loss-of-function screening in human organoids and identified 25 microcephaly-associated genes. In the aspect of treatment, the rescue of pathological phenotype in gene-corrected brain organoids provided a useful foundation for future research. Moreover, brain organoids should be used in high-throughput drug screening for congenital brain malformations. The creation of cell banks of hPSCs may provide a powerful resource for studies in this field and will enable studies to examine a larger cohort of patients (74).
Conclusions
The emergence of organoid technologies has opened new avenues for the recapitulation of human brain development, especially early neurodevelopment, and revealed the pathogenesis of congenital brain malformations. The accumulating advances enable increasingly accurate diagnoses and the development of personalized, targeted therapy for patients with congenital brain diseases. Several hurdles still need to be overcome, and future improvements will focus on combinations of in vitro vascularization and cutting-edge technologies.
Acknowledgments
We are grateful to Dr. Yiting Cai, from Children’s Hospital of Fudan University, for her careful proofreading of our paper. We also thank Figdraw (www.figdraw.com) and Servier Medical Art (smart.servier.com) for the assistance in creating the figures.
Funding: None.
Footnote
Reporting Checklist: The authors have completed the Narrative Review reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-22-239/rc
Peer Review File: Available at https://tp.amegroups.com/article/view/10.21037/tp-22-239/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-22-239/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chiaradia I, Lancaster MA. Brain organoids for the study of human neurobiology at the interface of in vitro and in vivo. Nat Neurosci 2020;23:1496-508. [Crossref] [PubMed]
- Buchsbaum IY, Kielkowski P, Giorgio G, et al. ECE2 regulates neurogenesis and neuronal migration during human cortical development. EMBO Rep 2020;21:e48204. [Crossref] [PubMed]
- Mora-Bermúdez F, Huttner WB. What Are the Human-Specific Aspects of Neocortex Development? Front Neurosci 2022;16:878950. [Crossref] [PubMed]
- Llorca A, Deogracias R. Origin, Development, and Synaptogenesis of Cortical Interneurons. Front Neurosci 2022;16:929469. [Crossref] [PubMed]
- Yang T, Veling MW, Zhao XF, et al. Migrating pyramidal neurons require DSCAM to bypass the border of the developing cortical plate. J Neurosci 2022;42:5510-21. [Crossref] [PubMed]
- Chaudhari BP, Ho ML. Congenital Brain Malformations: An Integrated Diagnostic Approach. Semin Pediatr Neurol 2022;42:100973. [Crossref] [PubMed]
- Zhang W, Ma L, Yang M, et al. Cerebral organoid and mouse models reveal a RAB39b-PI3K-mTOR pathway-dependent dysregulation of cortical development leading to macrocephaly/autism phenotypes. Genes Dev 2020;34:580-97. [Crossref] [PubMed]
- Hodge RD, Bakken TE, Miller JA, et al. Conserved cell types with divergent features in human versus mouse cortex. Nature 2019;573:61-8. [Crossref] [PubMed]
- Qian X, Song H, Ming GL. Brain organoids: advances, applications and challenges. Development 2019;146:dev166074. [Crossref] [PubMed]
- Tian A, Muffat J, Li Y. Studying Human Neurodevelopment and Diseases Using 3D Brain Organoids. J Neurosci 2020;40:1186-93. [Crossref] [PubMed]
- Eiraku M, Watanabe K, Matsuo-Takasaki M, et al. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell 2008;3:519-32. [Crossref] [PubMed]
- Eiraku M, Takata N, Ishibashi H, et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 2011;472:51-6. [Crossref] [PubMed]
- Lancaster MA, Renner M, Martin CA, et al. Cerebral organoids model human brain development and microcephaly. Nature 2013;501:373-9. [Crossref] [PubMed]
- Xiang Y, Cakir B, Park IH. Deconstructing and reconstructing the human brain with regionally specified brain organoids. Semin Cell Dev Biol 2021;111:40-51. [Crossref] [PubMed]
- Xiang Y, Cakir B, Park IH. Generation of Regionally Specified Human Brain Organoids Resembling Thalamus Development. STAR Protoc 2020;1:100001. [Crossref] [PubMed]
- Xiang Y, Tanaka Y, Patterson B, et al. Fusion of Regionally Specified hPSC-Derived Organoids Models Human Brain Development and Interneuron Migration. Cell Stem Cell 2017;21:383-398.e7. [Crossref] [PubMed]
- Cakir B, Xiang Y, Tanaka Y, et al. Engineering of human brain organoids with a functional vascular-like system. Nat Methods 2019;16:1169-75. [Crossref] [PubMed]
- Pham MT, Pollock KM, Rose MD, et al. Generation of human vascularized brain organoids. Neuroreport 2018;29:588-93. [Crossref] [PubMed]
- Gabriel E, Wason A, Ramani A, et al. CPAP promotes timely cilium disassembly to maintain neural progenitor pool. EMBO J 2016;35:803-19. [Crossref] [PubMed]
- Li R, Sun L, Fang A, et al. Recapitulating cortical development with organoid culture in vitro and modeling abnormal spindle-like (ASPM related primary) microcephaly disease. Protein Cell 2017;8:823-33. [Crossref] [PubMed]
- Zhang W, Yang SL, Yang M, et al. Modeling microcephaly with cerebral organoids reveals a WDR62-CEP170-KIF2A pathway promoting cilium disassembly in neural progenitors. Nat Commun 2019;10:2612. [Crossref] [PubMed]
- Omer Javed A, Li Y, Muffat J, et al. Microcephaly Modeling of Kinetochore Mutation Reveals a Brain-Specific Phenotype. Cell Rep 2018;25:368-382.e5. [Crossref] [PubMed]
- Esk C, Lindenhofer D, Haendeler S, et al. A human tissue screen identifies a regulator of ER secretion as a brain-size determinant. Science 2020;370:935-41. [Crossref] [PubMed]
- Dhaliwal N, Choi WWY, Muffat J, et al. Modeling PTEN overexpression-induced microcephaly in human brain organoids. Mol Brain 2021;14:131. [Crossref] [PubMed]
- Urresti J, Zhang P, Moran-Losada P, et al. Cortical organoids model early brain development disrupted by 16p11.2 copy number variants in autism. Mol Psychiatry 2021;26:7560-80. [Crossref] [PubMed]
- Fair SR, Schwind W, Julian D, et al. Cerebral organoids containing an AUTS2 missense variant model microcephaly. Brain 2023;146:387-404. [Crossref] [PubMed]
- Mariani J, Coppola G, Zhang P, et al. FOXG1-Dependent Dysregulation of GABA/Glutamate Neuron Differentiation in Autism Spectrum Disorders. Cell 2015;162:375-90. [Crossref] [PubMed]
- Li Y, Muffat J, Omer A, et al. Induction of Expansion and Folding in Human Cerebral Organoids. Cell Stem Cell 2017;20:385-396.e3. [Crossref] [PubMed]
- Villa CE, Cheroni C, Dotter CP, et al. CHD8 haploinsufficiency links autism to transient alterations in excitatory and inhibitory trajectories. Cell Rep 2022;39:110615. [Crossref] [PubMed]
- Dang LT, Vaid S, Lin G, et al. STRADA-mutant human cortical organoids model megalencephaly and exhibit delayed neuronal differentiation. Dev Neurobiol 2021;81:696-709. [Crossref] [PubMed]
- Bershteyn M, Nowakowski TJ, Pollen AA, et al. Human iPSC-Derived Cerebral Organoids Model Cellular Features of Lissencephaly and Reveal Prolonged Mitosis of Outer Radial Glia. Cell Stem Cell 2017;20:435-449.e4. [Crossref] [PubMed]
- Iefremova V, Manikakis G, Krefft O, et al. An Organoid-Based Model of Cortical Development Identifies Non-Cell-Autonomous Defects in Wnt Signaling Contributing to Miller-Dieker Syndrome. Cell Rep 2017;19:50-9. [Crossref] [PubMed]
- Jin M, Pomp O, Shinoda T, et al. Katanin p80, NuMA and cytoplasmic dynein cooperate to control microtubule dynamics. Sci Rep 2017;7:39902. [Crossref] [PubMed]
- O'Neill AC, Kyrousi C, Klaus J, et al. A Primate-Specific Isoform of PLEKHG6 Regulates Neurogenesis and Neuronal Migration. Cell Rep 2018;25:2729-2741.e6. [Crossref] [PubMed]
- Klaus J, Kanton S, Kyrousi C, et al. Altered neuronal migratory trajectories in human cerebral organoids derived from individuals with neuronal heterotopia. Nat Med 2019;25:561-8. [Crossref] [PubMed]
- Ayo-Martin AC, Kyrousi C, Di Giaimo R, et al. GNG5 Controls the Number of Apical and Basal Progenitors and Alters Neuronal Migration During Cortical Development. Front Mol Biosci 2020;7:578137. [Crossref] [PubMed]
- Pirozzi F, Nelson B, Mirzaa G. From microcephaly to megalencephaly: determinants of brain size. Dialogues Clin Neurosci 2018;20:267-82. [Crossref] [PubMed]
- Gabriel E, Ramani A, Altinisik N, et al. Human Brain Organoids to Decode Mechanisms of Microcephaly. Front Cell Neurosci 2020;14:115. [Crossref] [PubMed]
- Herber S, Silva AA, Sanseverino MTV, et al. Prevalence and causes of congenital microcephaly in the absence of a Zika virus outbreak in southern Brazil. J Pediatr (Rio J) 2019;95:600-6. [Crossref] [PubMed]
- Haffner DN, O'Connor S, Zempel J. A Rare Presentation of Congenital TORCH Infection. Pediatr Neurol 2020;105:71-2. [Crossref] [PubMed]
- Garcez PP, Loiola EC, Madeiro da Costa R, et al. Zika virus impairs growth in human neurospheres and brain organoids. Science 2016;352:816-8. [Crossref] [PubMed]
- Cugola FR, Fernandes IR, Russo FB, et al. The Brazilian Zika virus strain causes birth defects in experimental models. Nature 2016;534:267-71. [Crossref] [PubMed]
- Krenn V, Bosone C, Burkard TR, et al. Organoid modeling of Zika and herpes simplex virus 1 infections reveals virus-specific responses leading to microcephaly. Cell Stem Cell 2021;28:1362-1379.e7. [Crossref] [PubMed]
- Xu M, Lee EM, Wen Z, et al. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat Med 2016;22:1101-7. [Crossref] [PubMed]
- Li C, Deng YQ, Wang S, et al. 25-Hydroxycholesterol Protects Host against Zika Virus Infection and Its Associated Microcephaly in a Mouse Model. Immunity 2017;46:446-56. [Crossref] [PubMed]
- Zhou T, Tan L, Cederquist GY, et al. High-Content Screening in hPSC-Neural Progenitors Identifies Drug Candidates that Inhibit Zika Virus Infection in Fetal-like Organoids and Adult Brain. Cell Stem Cell 2017;21:274-283.e5. [Crossref] [PubMed]
- Pettke A, Tampere M, Pronk R, et al. Broadly Active Antiviral Compounds Disturb Zika Virus Progeny Release Rescuing Virus-Induced Toxicity in Brain Organoids. Viruses 2020;13:37. [Crossref] [PubMed]
- Cavalcante BRR, Aragão-França LS, Sampaio GLA, et al. Betulinic Acid Exerts Cytoprotective Activity on Zika Virus-Infected Neural Progenitor Cells. Front Cell Infect Microbiol 2020;10:558324. [Crossref] [PubMed]
- Pelletier F, Perrier S, Cayami FK, et al. Endocrine and Growth Abnormalities in 4H Leukodystrophy Caused by Variants in POLR3A, POLR3B, and POLR1C. J Clin Endocrinol Metab 2021;106:e660-74. [Crossref] [PubMed]
- Jones S, Samanta D. Macrocephaly. StatPearls. Treasure Island (FL), 2021.
- Subramanian L, Bershteyn M, Paredes MF, et al. Dynamic behaviour of human neuroepithelial cells in the developing forebrain. Nat Commun 2017;8:14167. [Crossref] [PubMed]
- Andrews MG, Subramanian L, Kriegstein AR. mTOR signaling regulates the morphology and migration of outer radial glia in developing human cortex. Elife 2020;9:e58737. [Crossref] [PubMed]
- Di Donato N, Chiari S, Mirzaa GM, et al. Lissencephaly: Expanded imaging and clinical classification. Am J Med Genet A 2017;173:1473-88. [Crossref] [PubMed]
- Juric-Sekhar G, Hevner RF. Malformations of Cerebral Cortex Development: Molecules and Mechanisms. Annu Rev Pathol 2019;14:293-318. [Crossref] [PubMed]
- Karzbrun E, Kshirsagar A, Cohen SR, et al. Human Brain Organoids on a Chip Reveal the Physics of Folding. Nat Phys 2018;14:515-22. [Crossref] [PubMed]
- Scott G, Huang Y. Engineering cerebral folding in brain organoids. Neural Regen Res 2022;17:2420-2. [Crossref] [PubMed]
- Rothenbücher TSP, Gürbüz H, Pereira MP, et al. Next generation human brain models: engineered flat brain organoids featuring gyrification. Biofabrication 2021;13:011001. [Crossref] [PubMed]
- Lancaster MA. Crinkle-Cut Brain Organoids. Cell Stem Cell 2018;22:616-8. [Crossref] [PubMed]
- Lewitus E, Kelava I, Kalinka AT, et al. Comment on "Cortical folding scales universally with surface area and thickness, not number of neurons". Science 2016;351:825. [Crossref] [PubMed]
- Mota B, Herculano-Houzel S. BRAIN STRUCTURE. Cortical folding scales universally with surface area and thickness, not number of neurons. Science 2015;349:74-7. [Crossref] [PubMed]
- Heinzen EL, O'Neill AC, Zhu X, et al. De novo and inherited private variants in MAP1B in periventricular nodular heterotopia. PLoS Genet 2018;14:e1007281. [Crossref] [PubMed]
- Giandomenico SL, Sutcliffe M, Lancaster MA. Generation and long-term culture of advanced cerebral organoids for studying later stages of neural development. Nat Protoc 2021;16:579-602. [Crossref] [PubMed]
- Giandomenico SL, Mierau SB, Gibbons GM, et al. Cerebral organoids at the air-liquid interface generate diverse nerve tracts with functional output. Nat Neurosci 2019;22:669-79. [Crossref] [PubMed]
- Lancaster MA, Corsini NS, Wolfinger S, et al. Guided self-organization and cortical plate formation in human brain organoids. Nat Biotechnol 2017;35:659-66. [Crossref] [PubMed]
- Mansour AA, Gonçalves JT, Bloyd CW, et al. An in vivo model of functional and vascularized human brain organoids. Nat Biotechnol 2018;36:432-41. [Crossref] [PubMed]
- Abud EM, Ramirez RN, Martinez ES, et al. iPSC-Derived Human Microglia-like Cells to Study Neurological Diseases. Neuron 2017;94:278-293.e9. [Crossref] [PubMed]
- Sloan SA, Darmanis S, Huber N, et al. Human Astrocyte Maturation Captured in 3D Cerebral Cortical Spheroids Derived from Pluripotent Stem Cells. Neuron 2017;95:779-790.e6. [Crossref] [PubMed]
- Madhavan M, Nevin ZS, Shick HE, et al. Induction of myelinating oligodendrocytes in human cortical spheroids. Nat Methods 2018;15:700-6. [Crossref] [PubMed]
- Kanton S, Treutlein B, Camp JG. Single-cell genomic analysis of human cerebral organoids. Methods Cell Biol 2020;159:229-56. [Crossref] [PubMed]
- Pollen AA, Bhaduri A, Andrews MG, et al. Establishing Cerebral Organoids as Models of Human-Specific Brain Evolution. Cell 2019;176:743-756.e17. [Crossref] [PubMed]
- Velasco S, Kedaigle AJ, Simmons SK, et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature 2019;570:523-7. [Crossref] [PubMed]
- Qian X, Nguyen HN, Song MM, et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 2016;165:1238-54. [Crossref] [PubMed]
- Renner H, Grabos M, Becker KJ, et al. A fully automated high-throughput workflow for 3D-based chemical screening in human midbrain organoids. Elife 2020;9:e52904. [Crossref] [PubMed]
- Li M, Zhao H, Ananiev GE, et al. Establishment of Reporter Lines for Detecting Fragile X Mental Retardation (FMR1) Gene Reactivation in Human Neural Cells. Stem Cells 2017;35:158-69. [Crossref] [PubMed]


