
Identification of cystic fibrosis transmembrane conductance regulator as a prognostic marker for juvenile myelomonocytic leukemia via the whole-genome bisulfite sequencing of monozygotic twins and data mining
Introduction
Juvenile myelomonocytic leukemia (JMML) is a unique myelodysplastic/myeloproliferative neoplasm caused by the excessive proliferation of monocytes and granulocytes in infancy/early childhood (1). Its clinical manifestations include hepatosplenomegaly, lymphadenopathy, skin rash, leukocytosis, monocytosis, thrombocytopenia, anemia, and respiratory failure. With a global annual incidence of 1.2/1,000,000, JMML accounts for 2–3% of all hematological malignancies. The median age of JMML patients at the time of diagnosis is 2 years, and the male-to-female ratio is 2–3 to 1 (2). Allogeneic hematopoietic stem cell transplantation (allo-HSCT) remains the only cure for JMML. However, the low 5-year overall survival rate of 50–60% and the high post-HSCT recurrence rate remain major challenges for allo-HSCT (3). Recurrence is still the main reason for treatment failure in JMML patients.
Approximately 90% of JMML patients harbor germline or somatic mutations in Protein Tyrosine Phosphatase Non-Receptor Type 11 (PTPN11), NRAS Proto-Oncogene, GTPase (NRAS), KRAS Proto-Oncogene, GTPase (KRAS), Cbl Proto-Oncogene (CBL), or Neurofibromin 1 (NF1) (4). The mutation of these genes can activate the RAS signaling pathway (5). Epigenetics is the study of heritable alterations in gene expression that are not due to changes in the deoxyribonucleic acid (DNA) sequence. Epigenetic modifications alter DNA accessibility and/or the chromatin structure by which the expression levels of certain genes are regulated (6). DNA methylation (DNAm) involves the addition of a methyl group at position C5 in the DNA cytosine ring catalyzed by DNA methyltransferase enzymes (7). As an epigenetic mechanism involving the transfer of a methyl group onto the C5 position of the cytosine to form 5-methylcytosine-, the DNAm patterns observed in cancer genomes can be classified as either gene promoter hypormethylated and hypermethylated. CpG islands (CGIs) located in promoter regions in normal human tissues are in a hypomethylated state. However, the promoter hypermethylation of tumor suppressor genes can silence those genes and cause cancer (8). Thus, DNAm is an important aspect to consider when studying human diseases. Whole-genome bisulfite sequencing (WGBS) can detect the precise boundaries between methylated and unmethylated regions at single-base resolution and is considered the gold-standard method for DNAm analysis (9).
A study showed that some genes are hypermethylated or have superhigh methylation in JMML patients. This study also reported 4 hypermethylated genes (i.e., BMP4, CALCA, CDKN2B, and RARB) (10). Previous studies have shown an association between activation of the DNA methylation machinery and specific JMML mutational profiles (11). Moreover, numerous studies have suggested that aberrant DNAm is linked to poor outcomes in JMML (10,12,13). However, clinical outcomes were significantly improved by treatment with the DNA-hypomethylating agent azacitidine prior to HSCT in patients with JMML (14). Thus, DNA hypermethylation is a novel independent prognostic factor for the poor prognosis of JMML (15). Identifying promising key genes associated with the progression and prognosis of JMML could lead to a marked improvement in the long-term survival of patients. Previous research has mainly focused on quantitative measurements of DNAm in large cohorts using the Infinium Human Methylation 450 Bead Chip (Illumina), quantitative high-resolution mass spectrometry (Sequenom MassARRAY), or bisulfite conversion and pyrosequencing techniques (13,16). To the best of the authors’ knowledge, the relationship between gene methylation and the carcinogenesis of JMML has not been reported. Monozygotic (MZ) twins essentially share identical genomes and early-childhood family environments. An MZ twin study is thus an ideal approach for examining the effects of epigenetic modifications on various diseases. In this study, we conducted the WGBS of twins, a JMML patient and his healthy brother, and data mined from a Gene Expression Omnibus (GEO) data set to explore the causative genes and their potential effects on JMML. We present the following article in accordance with the MDAR reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-22-381/rc).
Methods
General information about the patient
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Medical Ethics Committees of Nanfang Hospital, Southern Medical University (No. NFEC-2020-203). Before participating in this study, the patient’s parents provided written informed consent. The patient was a 6-year-old boy diagnosed with JMML. On first admission, his white blood cell count was 42.79×109/L, his monocyte count was 13.48×109/L, his platelet count was 21×109/L, and his hemoglobin was 21 g/dL. The examination showed that the bone marrow of the patient contained 8% naive granulocytes and 7% promyelocytes. The cytogenetic analysis showed a normal karyotype (46, XY), while targeted next-generation sequencing revealed an NF1 (S1399fs/R1748X) mutation. A physical examination showed splenomegaly, with a mean spleen size of 7 cm under the costal margin, and hepatomegaly, with a mean liver size of 2 cm under the costal margin. The patient appeared pale and had many café-au-lait spots on his lower limbs and trunk. His older brother was in good health.
Sample preparation
Bone marrow samples of the patient and peripheral blood samples of his older sibling and a healthy donor were collected. Total DNA from the above-mentioned samples was extracted using DNA extraction kits (TIANGEN, Beijing, China) following the manufacturer’s protocol.
WGBS
Isolated genomic DNA was sheared into fragments of 100–300 bp in length. The 3' and 5' overhangs were repaired to create blunt ends, and a single “A” was added to the 3' end of the blunt fragment. Methylated adapters were ligated to the A-tailed fragment and bisulfite modified using a ZYMO EZ DNA Methylation-Gold Kit. A final polymerase chain reaction (PCR) amplification step was then performed for library qualification. Only the qualified library with the targeted size range was sent for sequencing.
Genome-wide methylation analysis and gene set enrichment analysis
Global DNAm profiles in all 3 samples were examined. The data were then analyzed by a pairwise comparison to identify the key genes. DNA methylation in eukaryotes occurs on cytosine bases in the context of CG, CHG, and CHH (H = A, C, or T). First, the CG, CHG, and CHH methylation in each sample were analyzed separately to compare the DNAm features. Next, the average DNAm levels of 7 different transcription elements in the genome, including the upstream region, first exon, first intron, internal exon, internal intron, last exon, and downstream region, were analyzed. Next, the differentially methylated regions (DMRs) among the samples were analyzed and compared. We performed a Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis and a Gene Ontology (GO) enrichment analysis of the DMR-related genes to explore the role of epigenetic variations in pathways and biological processes and identified the overlapping differential signaling pathways. Network an enrichment analysis of the overlapping genes was performed using Cytoscape and Search Tool for the Retrieval of Interacting Genes/Proteins (STRING), which are gene/protein interaction databases, to identify the hub genes.
Real-time qPCR
The total ribonucleic acid (RNA) was extracted using TRIzol reagent (Invitrogen) from bone marrow blood samples from 3 JMML patients and peripheral blood samples from 3 healthy donors, and complementary DNA (cDNA) was reverse transcribed using a cDNA synthesis kit (Yeasen, Shanghai, China). The quantitative gene expression analysis was performed in an ABI7500 Real-Time PCR System (Applied Biosystems, California, USA) with a quantitative PCR (qPCR) SYBR Green Master Mix Kit (Yeasen, Shanghai, China). β-actin was used as the housekeeping gene, and the 2−ΔΔCt method was used to quantify the relative expression levels of the genes of interest. The primers used to amplify cystic fibrosis transmembrane conductance regulator (CFTR) and β-actin are listed in Table S1.
Data mining and survival analysis
A data mining platform was used to query the expression level data for the hub genes identified in the healthy controls and the JMML patient samples in the GEO database, and differences between the means were compared by using a 2-tailed Student’s t-test. For the survival analysis, the samples were divided into 2 groups according to the median expression levels of the hub genes, and the overall survival rates of the 2 groups were compared. A result was deemed to be statistically significant if the P value was <0.05.
Results
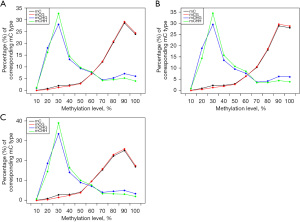
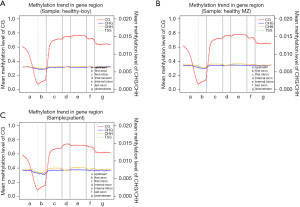
The analysis showed that the CHG and CHH methylation levels in the 3 children were similar. However, the distribution of cytosine methylation (mC) and CG methylation (mCG) in the JMML patients differed significantly between the healthy sibling and the healthy control (see Figure 1). We also analyzed the average DNAm levels of different transcription elements in the genomes of the 3 subjects and found no significant differences (see Figure 2).
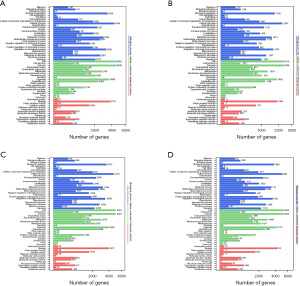
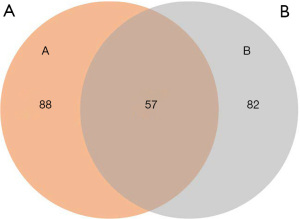
A KEGG pathway enrichment analysis and GO functional annotation analysis were performed on the DMR-associated genes in the 3 subjects. The DMR-related genes in the subjects had high consistency in functional clustering (P>0.05; see Figure 3). Interestingly, we found no significant differences between the heathy controls and the patient in the genes, but the results of KEGG pathway enrichment analysis showed significant differences in multiple signaling pathways in the promoter region (P<0.01; see Figure 4 and https://cdn.amegroups.cn/static/public/tp-22-381-1.xlsx, https://cdn.amegroups.cn/static/public/tp-22-381-2.xlsx). As the Venn diagram analysis shows, 57 pathways, including the RAS signaling pathway and the pathways associated with cancer, were significantly enriched and overlapped in the 3 subjects (see Figure 5 and https://cdn.amegroups.cn/static/public/tp-22-381-3.xlsx).

We performed a gene set enrichment analysis followed by network visualization of the gene data in the 57 overlapping pathways using the Cytoscape and STRING databases. The nodes represent the indicated genes, and the colored nodes represent the genes enriched in certain signaling pathways. The gene set enrichment analysis revealed that the node in red (i.e., CFTR) was the hub gene (see Figure 6).
qPCR was used for validation
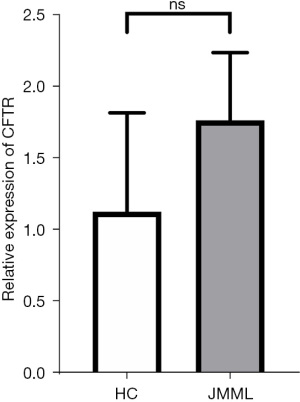
The relative expression levels of the target genes were verified in the 3 selected JMML patients and 3 healthy children’s bone marrow or peripheral blood. The leukemia group showed slightly higher expression levels of CFTR; however, the difference was not statistically significant (P=0.22; see Figure 7).
Data mining and statistical analysis
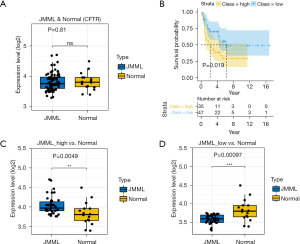
Publicly available data from the GEO database (https://www.ncbi.nlm.nih.gov/gds/) were mined. We removed batch effects between the 2 cohorts using the ComBat function in the sva package (17). The mean values between the patients and controls were compared using 2-tailed t-tests. As the box plot shows (see Figure 8A), the CFTR expression levels did not differ significantly between the JMML patients and healthy donors (GEO: GSE71935–JMML =38, normal =9; GEO: GSE71449–JMML =44, normal =7; P=0.81). However, according to the median gene expression levels in the microarray analysis, we divided patients into low- and high-expression subgroups. The Kaplan-Meier curve for the 82 well-defined JMML patients enrolled in the database revealed that higher CFTR expression was linked to lower survival probability (P=0.019; see Figure 8B). A comparison of the different prognosis subgroups of the JMML patients with the healthy controls showed that the expression of CFTR was significantly elevated in the poor prognosis group (P=0.0049; see Figure 8C). Conversely, patients with a lower expression level of CFTR may had a relatively better prognosis (P=0.00097; see Figure 8D).
Discussion
JMML is a clonal myeloproliferative/myelodysplastic neoplasia. Approximately 35% of JMML patients who receive HSCT relapse within 5 years, indicating the strong need for a better understanding of the molecular mechanisms of JMML (3). Our study identified the important role of DNAm in the development and progression of JMML. A total of 57 overlapping signaling pathways, including RAS, were identified as target pathways that were involved in the pathogenesis of JMML. Some studies have described candidate genes that are transcriptionally regulated by methylation in JMML (10,12); however, this study is the first to show that CFTR gene methylation is significantly correlated with JMML. A total of 5 signaling pathways (i.e., the Cyclic Adenosine monophosphate (cAMP) signaling, AMP-activated protein kinase (AMPK) signaling, bile secretion, gastric acid secretion, and pancreatic secretion pathways) were identified in this study (see https://cdn.amegroups.cn/static/public/tp-22-381-4.xlsx). Due to the limited sample size, no significant difference between the JMML patients and healthy children was found in relation to the expression of CFTR. However, a survival analysis of a larger cohort indicated that patients with high expression levels of CFTR may exhibit a poorer prognosis than patients with low expression levels of CFTR.
CFTR, which is critical for carcinogenesis, was identified as one of the hub genes (18). CFTR is a glycoprotein with 1480 amino acids that belongs to the family of Adenosine triphosphate binding cassette (ABC) transporters (15). ABC transporters are frequently overexpressed in metastatic cancers, contributing to chemoresistance (19). Additionally, CFTR is a cAMP-regulated chloride channel that contains 2 nucleotide-binding domains (NBDs) (i.e., NBD1, and NBD2), and a cytosolic region called the R domain in addition to 12 transmembrane helices (15). A function of CFTR is to transport Cl− and HCO3−, and it also regulates other ion channels (Na+, K+, Ca2+, and other Cl− channels) (20-22). Additionally, CFTR also has roles in osmoregulation, membrane potential maintenance, lipid homeostasis, cell polarity, the metabolism of glucose and other substrates, oxidative stress, inflammation, mucus production, microbiome alterations, pH regulation, cell motility, autophagy, mitochondrial dysfunction, apoptosis, cell polarity, cell-cell contact, stem cell function, and cellular immune responses (23-33). Some studies have shown multiple associations between CFTR and cancer; however, its expression levels vary between different types of tumors. DNAm is the major epigenetic approach for gene regulation, and the destruction of DNAm is related to a variety of diseases (34). The downregulation of the CFTR gene by promoter methylation has been demonstrated in various cancer types, including lung cancer (35), liver cancer (36), and head and neck cancer (37). However, CFTR is overexpressed in other cancers, such as ovarian cancer (38). This discrepancy indicates that the CFTR gene may act as both a proto-oncogene and an anti-oncogene.
The dual role of CFTR may be linked to its complex gene expression pattern, and the interaction of its promoter with intronic enhancers may coordinate gene transcription (39,40). In addition to being expressed in actively proliferating epithelial cells, CFTR is also widely expressed in immune cells of the blood system and exerts biological functions, and its anion transport and regulation characteristics are the same as those of epithelial cells (41,42). The epigenetic regulation of CFTR has been discovered in other solid tumors, but very few studies of hematologic malignancies have been reported. Indeed, until 2017, only 1 study had shown that CFTR acts as an oncogene in Ph+ acute lymphoblastic leukemia (Ph+ALL) (43), and the expression level of the CFTR in Ph+ALL patients was found to be higher than that of healthy controls in another study (44). However, the relationship between CFTR and the development of JMML remains unclear.
In this study, consistent with similar results for other solid tumors, we found that the promoter of CFTR was hypermethylated in JMML patients. The CFTR promoter is a “housekeeping” type promoter rich in CpG. In addition to promoter methylation, hypoxia-responsive elements, intronic enhancers, and insulator elements that functionally interact with promoters in a cell-type-specific manner can also partially control the spatial regulation of CpG CFTR expression (45). Generally, DNA promoter methylation and gene expression are negatively correlated, but the high expression levels of some genes can be maintained even when their promoter regions are methylated (46). Further, promoter methylation status has been shown to be correlated with prognosis; for example, the hypermethylation of CFTR was found to be associated with an unfavorable survival rate in patients with prostate cancer (47).
Based on the role of CFTR in other tumors and the results of our methylation analysis, it is reasonable to assume that CFTR is an important JMML-associated gene. Thus, we downloaded microarray data sets and clinical follow-up data from the GEO database. The Kaplan-Meier curves revealed that high CFTR expression was associated with inferior prognosis in JMML, suggesting that CFTR and JMML may have a close relationship. However, this possibility requires further exploration.
Autophagy has emerged as an effective escape mechanism for promoting tolerance to chemotherapeutic drugs, ultimately leading to poor clinical outcomes (48). The constitutive activation of the RAS signaling pathway, the main pathogenic mechanism of JMML, is closely related to autophagy. Inhibition of KRAS→RAF→MEK→ERK signaling triggers autophagy, protecting cancer cells from the cytotoxic effects of RAS signaling pathway inhibition (49). However, high expression of CFTR may lead to autophagy and resistance to chemotherapeutic drugs in cancer cells (50). In terms of treatment efficiency, there is currently no type of chemotherapy that can lead to long-term remission in JMML patients.
Studies have shown that CFTR expression is also upregulated in ovarian cancer (37). The Ras-MAPK/Erk-ETS1/CFTR1 axis was found to be upregulated in ovarian cancer cell lines. This upregulation may increase the proliferation and invasion and reduce the drug absorption and apoptosis of tumor cells, ultimately resulting in chemotherapeutic resistance (51). ETS1, a main downstream effector in the Ras/ERK signaling pathway, is generally considered a transcriptional activator and is commonly hyperactivated in cancer. More specifically, the Ras-MAPK signaling pathway can be activated by ETS1 via a dual functional role (52). Increased ETS1 expression in ovarian cancer was found to be associated with a poor prognosis (53), and ETS1 can regulate the expression of CFTR by binding to its promoter regions (50). These findings suggest that CFTR is associated with the ETS1 and Ras signaling pathways. Additionally, the overexpression of CFTR in serous ovarian cancer can activate the c-Src signaling pathways (54), which cooperate with the RAS signaling pathway to promote tumorigenesis (55). Despite this evidence, more studies need to be conducted to demonstrate the direct relationship between the overexpression of CFTR and the activation of the RAS/ERK signaling pathway.
Conversely, the high expression of CFTR is positively correlated with nuclear factor kappa beta (NF-κB) activation (56). Activated NF-κB binds to the src homology 2 (SH2) domain-containing tyrosine phosphatase-2 (SHP2) promoter, leading to increased SHP2 expression, which enhances the activation of the RAS/RAF/MEK/ERK pathway (57). The constitutive activation of the RAS pathway is the main pathogenic mechanism of JMML (4,5). Conversely, microRNA -150-5p, a small non-coding RNA, was discovered to play roles in both cancer and autoimmune disease (58,59). CFTR-mediated HCO3− influx can activate soluble adenylate cyclase (sAC), which in turn activates protein kinase A (PKA)-dependent NF-κB signaling (60). Activated NF-κB1 interacts with miR-150-5p, which negatively regulates the miR-150-5p expression level (61). The downregulation of miR-150-5p leads to the phosphorylation of STAT5b and activates KRAS, NRAS, NF1, and PTPN11 in JMML (62). This observation suggests that microRNAs might function as intermediate links between CFTR and JMML pathogenesis/disease progression. Thus, we speculate that CFTR and the pathogenesis of JMML may be closely connected via RAS pathway regulation. Further investigations urgently need to be conducted to identify the mechanisms underlying this relationship. Subsequent research work will focus on genome-wide methylation analysis in expanded samples of JMML patients, and functional verification at the cellular level.
This was a preliminary laboratory study; thus, further investigation of CFTR function both in vivo and in vitro are required, and more investigations are needed to identify the mechanisms of CFTR in JMML development and progression. In future clinical research, to evaluate the relationship between CFTR promoter methylation and the clinical prognosis of JMML, we will increase the number of cases to dynamically monitor any changes in CFTR promoter methylation in JMML patients during treatment. Due to restrictions related to our sample size, our research lacked genome-wide methylation data and the corresponding gene expression profiling data from a large sample. In the next study, the sample size needs to be increased to elucidate this mechanism through a more thorough experimental analysis.
In conclusion, this study was the first to show that the promoter of CFTR is hypermethylated in JMML. This was a preliminary laboratory study but combined with the clinical data demonstrated in the study, we are of the view that CFTR could be a promising diagnostic, therapeutic, and prognostic biomarker for JMML. However, further investigations of CFTR function both in vivo and in vitro are required, and more investigations are needed to reveal the role of the CFTR gene in JMML development and progression. In future clinical research, to deepen the knowledge of the relationship between CFTR promoter methylation and the clinical prognosis of JMML, the authors will increase the sample size to dynamically monitor the changes in CFTR promoter methylation in JMML patients during treatment. Genome-wide methylation data of more cases will also be taken into consideration to elucidate the matter.
Acknowledgments
We would like to thank the subjects and their parents for participating in this study.
Funding: This study was supported by the Clinical Research Program of Nanfang Hospital, Southern Medical University (No. 2018CR042), the Clinical Research Startup Program of Southern Medical University by High-Level University Construction Funding of Guangdong Provincial Department of Education (No. LC2016ZD017), and the Science and Technology Planning Project of Guangdong Province of China (No. 2016A020215102, to XW).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-22-381/rc
Data Sharing Statement: Available at https://tp.amegroups.com/article/view/10.21037/tp-22-381/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-22-381/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by the Medical Ethics Committees of Nanfang Hospital, Southern Medical University (No. NFEC-2020-203). Before participating in this study, the patient’s parents provided written informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Hasle H, Kerndrup G, Jacobsen BB. Childhood myelodysplastic syndrome in Denmark: incidence and predisposing conditions. Leukemia 1995;9:1569-72. [PubMed]
- Loh ML. Childhood myelodysplastic syndrome: focus on the approach to diagnosis and treatment of juvenile myelomonocytic leukemia. Hematology Am Soc Hematol Educ Program 2010;2010:357-62. [Crossref] [PubMed]
- Locatelli F, Nöllke P, Zecca M, et al. Hematopoietic stem cell transplantation (HSCT) in children with juvenile myelomonocytic leukemia (JMML): results of the EWOG-MDS/EBMT trial. Blood 2005;105:410-9. [Crossref] [PubMed]
- Gupta AK, Meena JP, Chopra A, et al. Juvenile myelomonocytic leukemia-A comprehensive review and recent advances in management. Am J Blood Res 2021;11:1-21. [PubMed]
- Gazin C, Wajapeyee N, Gobeil S, et al. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature 2007;449:1073-7. [Crossref] [PubMed]
- Handy DE, Castro R, Loscalzo J. Epigenetic modifications: basic mechanisms and role in cardiovascular disease. Circulation 2011;123:2145-56. [Crossref] [PubMed]
- Allis CD. Epigenetics. New York, NY: CSHL Press, 2007.
- Weaver IC, Cervoni N, Champagne FA, et al. Epigenetic programming by maternal behavior. Nat Neurosci 2004;7:847-54. [Crossref] [PubMed]
- Cokus SJ, Feng S, Zhang X, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 2008;452:215-9. [Crossref] [PubMed]
- Olk-Batz C, Poetsch AR, Nöllke P, et al. Aberrant DNA methylation characterizes juvenile myelomonocytic leukemia with poor outcome. Blood 2011;117:4871-80. [Crossref] [PubMed]
- Lipka DB., Witte T, Toth R, et al. RAS-pathway mutation patterns define epigenetic subclasses in juvenile myelomonocytic leukemia. Nat. Commun. 2017;8:2126. [Crossref] [PubMed]
- Wilhelm T, Lipka DB, Witte T, et al. Epigenetic silencing of AKAP12 in juvenile myelomonocytic leukemia. Epigenetics 2016;11:110-9. [Crossref] [PubMed]
- Sakaguchi H, Muramatsu H, Okuno Y, et al. Aberrant DNA Methylation Is Associated with a Poor Outcome in Juvenile Myelomonocytic Leukemia. PLoS One 2015;10:e0145394. [Crossref] [PubMed]
- Locatelli F, Niemeyer CM. How I treat juvenile myelomonocytic leukemia. Blood 2015;125:1083-90. [Crossref] [PubMed]
- Schönung M, Meyer J, Nöllke P, et al. International consensus definition of DNA methylation subgroups in juvenile myelomonocytic leukemia. Clin. Cancer Res. 2021;27:158. [Crossref] [PubMed]
- Lipka DB, Witte T, Toth R, et al. RAS-pathway mutation patterns define epigenetic subclasses in juvenile myelomonocytic leukemia. Nat Commun 2017;8:2126. [Crossref] [PubMed]
- Leek JT, Johnson WE, Parker HS, et al. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 2012;28:882-3. [Crossref] [PubMed]
- Guo Y, Xing Y. Weighted gene co-expression network analysis of pneumocytes under exposure to a carcinogenic dose of chloroprene. Life Sci 2016;151:339-47. [Crossref] [PubMed]
- He J, Fortunati E, Liu DX, et al. Pleiotropic Roles of ABC Transporters in Breast Cancer. Int J Mol Sci 2021;22:3199. [Crossref] [PubMed]
- Anderson MP, Gregory RJ, Thompson S, et al. Demonstration that CFTR is a chloride channel by alteration of its anion selectivity. Science 1991;253:202-5. [Crossref] [PubMed]
- Quinton PM. The neglected ion: HCO3-. Nat Med 2001;7:292-3. [Crossref] [PubMed]
- Anderson KJ, Cormier RT, Scott PM. Role of ion channels in gastrointestinal cancer. World J Gastroenterol 2019;25:5732-72. [Crossref] [PubMed]
- Prevarskaya N, Skryma R, Shuba Y. Ion Channels in Cancer: Are Cancer Hallmarks Oncochannelopathies? Physiol Rev 2018;98:559-621. [Crossref] [PubMed]
- Lastraioli E, Iorio J, Arcangeli A. Ion channel expression as promising cancer biomarker. Biochim Biophys Acta 2015;1848:2685-702. [Crossref] [PubMed]
- Pankonien I, Quaresma MC, Rodrigues CS, et al. CFTR, Cell Junctions and the Cytoskeleton. Int J Mol Sci 2022;23:2688. [Crossref] [PubMed]
- Keown K, Brown R, Doherty DF, et al. Airway Inflammation and Host Responses in the Era of CFTR Modulators. Int J Mol Sci 2020;21:6379. [Crossref] [PubMed]
- Vernocchi P, Del Chierico F, Russo A, et al. Gut microbiota signatures in cystic fibrosis: Loss of host CFTR function drives the microbiota enterophenotype. PLoS One 2018;13:e0208171. [Crossref] [PubMed]
- Kleme ML, Sané AT, Garofalo C, et al. Targeted CFTR gene disruption with zinc-finger nucleases in human intestinal epithelial cells induces oxidative stress and inflammation. Int J Biochem Cell Biol 2016;74:84-94. [Crossref] [PubMed]
- Kleme ML, Sané A, Garofalo C, et al. CFTR Deletion Confers Mitochondrial Dysfunction and Disrupts Lipid Homeostasis in Intestinal Epithelial Cells. Nutrients 2018;10:836. [Crossref] [PubMed]
- Xia X, Wang J, Liu Y, et al. Lower Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) Promotes the Proliferation and Migration of Endometrial Carcinoma. Med Sci Monit 2017;23:966-74. [Crossref] [PubMed]
- Liou TG. The Clinical Biology of Cystic Fibrosis Transmembrane Regulator Protein: Its Role and Function in Extrapulmonary Disease. Chest 2019;155:605-16. [Crossref] [PubMed]
- Villella VR, Venerando A, Cozza G, et al. A pathogenic role for cystic fibrosis transmembrane conductance regulator in celiac disease. EMBO J 2019;38:e100101. [Crossref] [PubMed]
- Gelfond D, Heltshe S, Ma C, et al. Impact of CFTR Modulation on Intestinal pH, Motility, and Clinical Outcomes in Patients With Cystic Fibrosis and the G551D Mutation. Clin Transl Gastroenterol 2017;8:e81. [Crossref] [PubMed]
- Egger G, Liang G, Aparicio A, et al. Epigenetics in human disease and prospects for epigenetic therapy. Nature 2004;429:457-63. [Crossref] [PubMed]
- Tian F, Zhao J, Fan X, et al. Weighted gene co-expression network analysis in identification of metastasis-related genes of lung squamous cell carcinoma based on the Cancer Genome Atlas database. J Thorac Dis 2017;9:42-53. [Crossref] [PubMed]
- Moribe T, Iizuka N, Miura T, et al. Methylation of multiple genes as molecular markers for diagnosis of a small, well-differentiated hepatocellular carcinoma. Int J Cancer 2009;125:388-97. [Crossref] [PubMed]
- Shin Y, Kim M, Won J, et al. Epigenetic Modification of CFTR in Head and Neck Cancer. J Clin Med 2020;9:734. [Crossref] [PubMed]
- Zhu LC, Hu ZH, Liu JJ, et al. Whole Genome Expression Profiling Analysis of Metastasis and Drug-resistance-related Genes in Epithelial Ovarian Cancer Cells. Zhongguo Yi Xue Ke Xue Yuan Xue Bao 2015;37:662-73. [PubMed]
- Ott CJ, Suszko M, Blackledge NP, et al. A complex intronic enhancer regulates expression of the CFTR gene by direct interaction with the promoter. J Cell Mol Med 2009;13:680-92. [Crossref] [PubMed]
- Lewandowska MA, Costa FF, Bischof JM, et al. Multiple mechanisms influence regulation of the cystic fibrosis transmembrane conductance regulator gene promoter. Am J Respir Cell Mol Biol 2010;43:334-41. [Crossref] [PubMed]
- Yoshimura K, Nakamura H, Trapnell BC, et al. Expression of the cystic fibrosis transmembrane conductance regulator gene in cells of non-epithelial origin. Nucleic Acids Res 1991;19:5417-23. [Crossref] [PubMed]
- Sun H, Wang Y, Zhang J, et al. CFTR mutation enhances Dishevelled degradation and results in impairment of Wnt-dependent hematopoiesis. Cell Death Dis 2018;9:275. [Crossref] [PubMed]
- Neviani P, Santhanam R, Trotta R, et al. The tumor suppressor PP2A is functionally inactivated in blast crisis CML through the inhibitory activity of the BCR/ABL-regulated SET protein. Cancer Cell 2005;8:355-68. [Crossref] [PubMed]
- Yang X, Yan T, Gong Y, et al. High CFTR expression in Philadelphia chromosome-positive acute leukemia protects and maintains continuous activation of BCR-ABL and related signaling pathways in combination with PP2A. Oncotarget 2017;8:24437-48. [Crossref] [PubMed]
- Gillen AE, Harris A. Transcriptional regulation of CFTR gene expression. Front Biosci (Elite Ed) 2012;4:587-92. [Crossref] [PubMed]
- Guillaumet-Adkins A, Richter J, Odero MD, et al. Hypermethylation of the alternative AWT1 promoter in hematological malignancies is a highly specific marker for acute myeloid leukemias despite high expression levels. J Hematol Oncol 2014;7:4. [Crossref] [PubMed]
- Ashour N, Angulo JC, Andrés G, et al. A DNA hypermethylation profile reveals new potential biomarkers for prostate cancer diagnosis and prognosis. Prostate 2014;74:1171-82. [Crossref] [PubMed]
- Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer 2017;17:528-42. [Crossref] [PubMed]
- Kinsey CG, Camolotto SA, Boespflug AM, et al. Protective autophagy elicited by RAF→MEK→ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat Med 2019;25:620-7. [Crossref] [PubMed]
- Zhu Q, Li H, Liu Y, et al. Knockdown of CFTR enhances sensitivity of prostate cancer cells to cisplatin via inhibition of autophagy. Neoplasma 2017;64:709-17. [Crossref] [PubMed]
- Rodriguez-Aguayo C, Bayraktar E, Ivan C, et al. PTGER3 induces ovary tumorigenesis and confers resistance to cisplatin therapy through up-regulation Ras-MAPK/Erk-ETS1-ELK1/CFTR1 axis. EBioMedicine 2019;40:290-304. [Crossref] [PubMed]
- Plotnik JP, Budka JA, Ferris MW, et al. ETS1 is a genome-wide effector of RAS/ERK signaling in epithelial cells. Nucleic Acids Res 2014;42:11928-40. [Crossref] [PubMed]
- Prasad P, Roy SS. Glutamine regulates ovarian cancer cell migration and invasion through ETS1. Heliyon 2021;7:e07064. [Crossref] [PubMed]
- Xu J, Lin L, Yong M, et al. Adenovirus-mediated overexpression of cystic fibrosis transmembrane conductance regulator enhances invasiveness and motility of serous ovarian cancer cells. Mol Med Rep 2016;13:265-72. [Crossref] [PubMed]
- Shields DJ, Murphy EA, Desgrosellier JS, et al. Oncogenic Ras/Src cooperativity in pancreatic neoplasia. Oncogene 2011;30:2123-34. [Crossref] [PubMed]
- Wu Z, Peng X, Li J, et al. Constitutive activation of nuclear factor κB contributes to cystic fibrosis transmembrane conductance regulator expression and promotes human cervical cancer progression and poor prognosis. Int J Gynecol Cancer 2013;23:906-15. [Crossref] [PubMed]
- Nichols RJ, Haderk F, Stahlhut C, et al. RAS nucleotide cycling underlies the SHP2 phosphatase dependence of mutant BRAF-, NF1- and RAS-driven cancers. Nat Cell Biol 2018;20:1064-73. [Crossref] [PubMed]
- Chang WA, Tsai MJ, Hung JY, et al. miR-150-5p-Containing Extracellular Vesicles Are a New Immunoregulator That Favor the Progression of Lung Cancer in Hypoxic Microenvironments by Altering the Phenotype of NK Cells. Cancers (Basel) 2021;13:6252. [Crossref] [PubMed]
- Ying W, Tseng A, Chang RC, et al. miR-150 regulates obesity-associated insulin resistance by controlling B cell functions. Sci Rep 2016;6:20176. [Crossref] [PubMed]
- Lu YC, Chen H, Fok KL, et al. CFTR mediates bicarbonate-dependent activation of miR-125b in preimplantation embryo development. Cell Res 2012;22:1453-66. [Crossref] [PubMed]
- Ma Y, Liu Y, Hou H, et al. MiR-150 predicts survival in patients with sepsis and inhibits LPS-induced inflammatory factors and apoptosis by targeting NF-κB1 in human umbilical vein endothelial cells. Biochem Biophys Res Commun 2018;500:828-37. [Crossref] [PubMed]
- Leoncini PP, Bertaina A, Papaioannou D, et al. MicroRNA fingerprints in juvenile myelomonocytic leukemia (JMML) identified miR-150-5p as a tumor suppressor and potential target for treatment. Oncotarget 2016;7:55395-408. [Crossref] [PubMed]


