
Anti-proliferation and apoptosis-inducing effects of dihydroartemisinin on SH-SY5Y cells and metabolomic analysis
Introduction
Neuroblastoma (NB) is a childhood cancer, and the median age at diagnosis is about 17 months. Although tumors can occur anywhere in the sympathetic nervous system, most occur in the adrenal medulla (1). High-risk NB patients have a poor prognosis, and survivors often have long-term sequelae due to treatment. The 5-year overall survival (OS) rate is only about 50% (2). Despite increasing knowledge about the occurrence of NB, there is still a lack of effective treatments (3). Therefore, new effective drugs are currently sought for NB treatment.
Since the Nobel Prize was awarded to Tu Youyou for their discovery of artemisinin (ART) in 2015, there has been an upsurge of research on ART and its derivatives. Emerging experimental evidence shows that, in addition to treating malaria, ART and its derivatives also have a wide range of effects in cancer, autoimmune diseases, and infectious diseases (4). Dihydroartemisinin (DHA), one of the semi-synthetic derivatives of ART, has been reported to inhibit the growth of prostate cancer, esophageal cancer, and cervical cancer (5-7) via regulating the intracellular redox state (8). Recent studies have further shown that DHA can induce the formation of oxygen free radicals and cell cycle arrest; decrease cell proliferation, cell invasion, and metastasis; and cause changes in tumor-related genes (9-11). A study has emerged on the anti-tumor activity of DHA (12), but it has been involved in limited clinical trials on NB; its clinical anticancer efficacy remains unknown. The effect of DHA on NB and its potential molecular mechanism are still unclear.
Apoptosis is a cellular self-destruction mechanism and is essential for a variety of biological events. Most anticancer drugs currently used in the clinical setting exploit the intact apoptotic signaling pathways to trigger cancer cell death (13). Reactive oxygen species (ROS) in cancer cells play a central role in regulating and inducing apoptosis, thereby modulating cancer cell proliferation, survival, and drug resistance (14). Recent research has found that DHA inhibits the proliferation of tumor cells via apoptosis and the upregulation of ROS generation in leukemia (15), pancreatic cancer (16), and colon cancer (17). These findings indicate that ROS and apoptosis may participate in the antitumor effect of DHA in human NB cells. Metabolomics is used to identify the set of metabolites that are associated with physiological conditions or aberrant processes (18). Metabolomics can more thoroughly search for drug targets by analyzing the final products of cellular regulatory pathways (19). High-throughput metabolomic data analysis will help deepen our understanding of the effect of DHA on NB.
We hypothesized that DHA may restrain cell proliferation and advance cell apoptosis in NB. Thus, the aim of this study was to identify the roles of DHA in cell proliferation and apoptosis in SH-SY5Y cells, and further explore its potential mechanisms. We reported that DHA induces ROS generation and increases poly (ADP-ribose) polymerase (PARP-1) and caspase 3 levels in SH-SY5Y cells. In addition, we used an untargeted ultra-high performance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS)-based metabolomic approach to identify altered metabolites. Therefore, in this study, we explained the preliminarily effect of DHA on SH-SY5Y cell metabolism and suggested a potential drug for the treatment of NB. We present the following article in accordance with the MDAR reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-22-331/rc).
Methods
Reagents
We purchased DHA, N-acetylcysteine (NAC), pifithrin-α (PFTα), and the cell cycle and apoptosis analysis kit from Meilun Biotech. (Dalian, China). Dichloro-dihydro-fluorescein diacetate (DCFH-DA) and the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay kit was purchased from Beyotime Biotechnology (Jiangsu, China).
Cell culture
We purchased the NB cells (SH-SY5Y) from Shanghai Cell Bank, Chinese Academy of Sciences (Shanghai, China). The SH-SY5Y cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM)/F12 medium containing 10% fetal bovine serum (FBS). Cells were cultured as a monolayer in 5% CO2 in a humidified incubator at 37 ℃.
Cell viability test
Cytotoxicity of DHA was detected using the MTT assay. The SH-SY5Y cells were adjusted to 4×105 cells/mL with DMEM/F12 medium; 100 µL cell suspension was seeded onto a 96-well plate. Various concentrations of DHA (0.675, 1.25, 2.5, 5, 10, 20, and 40 µM) were added into the plate and cultured for 24 hours. Then, 100 µL DMEM/F12 containing 0.5 mg/mL MTT solution was added to each well and incubated for 4 hours. Absorbance was measured at 490 nm to determine the cell viability.
Colony formation assay
The SH-SY5Y cells (2,000/well) were placed onto a 12-well plate. After incubation with 2 µM DHA for 10 days, the cells were fixed with 4% paraformaldehyde for 30 minutes and then stained with 0.1% crystal violet for 20 minutes.
Cell apoptosis analysis
Cells were cultured in serum-free medium for 12 hours to synchronize the cell cycle. Cells were seeded in 6-well plates at a density of 1×106 cells/well and cultured in medium containing 10% FBS. After 24 hours of treatment with DHA at the indicated concentration, the cells were harvested and washed with phosphate-buffered saline (PBS). Cell-cycle distribution and cell apoptosis were detected by the cell cycle and apoptosis analysis kit, according to the manufacturer’s protocol. Briefly, the cells were fixed with 70% ethanol and resuspended in PBS containing propidium iodide (PI; 50 mg/mL) and RNase A (10 mg/mL), and stained for 30 minutes in the dark. The percentage of apoptotic cells in single cell suspension was detected by flow cytometry (CytoFLEX; Beckman Coulter, Brea, CA, USA).
ROS detection
Cellular ROS was detected using DCFH-DA, according to the manufacturer’s protocol. In brief, SH-SY5Y cells were seeded onto a 6-well plate at the above density and treated with DHA for 24 hours. After the treatment, the cells were washed twice with PBS, labeled with 5 µM DCFH-DA for 30 minutes, and the fluorescence intensity of ROS was detected by a fluorescence microscope (Observer A1; Zeiss, Oberkochen, Germany).
Western blot
After the cells were treated with DHA, the supernatant was aspirated and washed twice with PBS on ice. Cells were lysed with radioimmunoprecipitation assay (RIPA) buffer containing protease and phosphatase inhibitors (Beyotime Biotechnology).
The protein concentration was quantified using the bicinchoninic acid (BCA) protein assay kit (Beyotime Biotechnology). In total, 20 µg protein was separated by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and then transferred onto a polyvinylidene difluoride (PVDF) membrane (Bio-Rad Laboratories, Hercules, CA, USA). After blocking with 5% skim milk, membranes were incubated with the indicated primary antibodies at 4 ℃ for 12 hours. After washing the PVDF membrane with tris-buffered saline with 0.1% Tween 20 (TBST), the membrane was incubated with a secondary antibody for 1 hour at room temperature. An enhanced chemiluminescence (ECL) detection system was used to measure the expression of target bands. The primary antibodies used in this experiment were as follows: anticleaved-PARP-1 [1:1,000, 9532; Cell Signaling Technology (CST), Danvers, MA, USA], anticleaved caspase 3 (1:1,000, AF5132; Beyotime), anti-GAPDH (1:2,000, 60004-1-Ig; Proteintech, Rosemont, IL, USA), and anti-γH2AX (1:1,000, ab22551; Abcam). The ImageJ software (National Institutes of Health, Bethesda, MD, USA) was used to analyze the gray value of the target band. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) served as the loading control.
Immunofluorescence assay
The SH-SY5Y cells were cultured on glass coverslips on a 24-well plate (2×105 cells/well) with or without DHA treatment. The samples were fixed, permeabilized, blocked, and incubated with γH2AX monoclonal antibody (1:500, ab22551; Abcam, Cambridge, MA, USA) at 4 ℃ for 12 hours, and then incubated with the corresponding secondary antibody at 37 ℃ for 1 hour. The cell nucleus was stained with 4',6-diamidino-2-phenylindole dihydrochloride (DAPI; 10 µg/mL) (Sigma Aldrich, St. Louis, MO, USA).
Cell sample preparation for metabolomics
After DHA treatment for 24 hours, 1×107 SH-SY5Y cells/well in different groups were collected. A total of 100 mg glass beads (Sigma) were added into a 2 mL EP tube. Then, 1,000 µL acetonitrile-methanol: aqueous solution (2:2:1, v/v/v) was added and vortexed for 30 seconds to mix the cells. The samples were then put in liquid nitrogen and frozen for 30 seconds, removed, and left to stand for 1 minute. The samples were placed into a grinder (TissueLyser II; QIAGEN, Hilden, Germany) for 1 minute at 60 Hz, and then centrifuged at 13,000× g for 10 minutes at 4 ℃. Then, 850–900 µL of supernatant was placed into a 2 mL centrifuge tube and concentrated to dryness with a vacuum concentrator (Concentrator plus; Eppendorf, Hamburg, Germany). Finally, 300 µL of acetonitrile: 0.1% formic acid (FA) (1:9, v/v) solution was added to reconstitute the sample. Cell samples were filtered through a 0.22-µm membrane for cell metabolomic analysis (20).
UPLC-MS/MS analysis
Samples were run on a Q Exactive mass spectrometer coupled to an UltiMate 3000 UHPLC system (Thermo Fisher Scientific, Waltham, MA, USA). A Waters ACQUITY UPLC HSS T3 1.8 µm (2.1 mm, 17×150 mm; Waters, Milford, MA, USA) was used for chromatographic separation. The column oven was set at 40 ℃, and the autosampler temperature was maintained at 8 ℃. Eluents C (water containing 0.1% FA) and D (acetonitrile containing 0.1% FA) were employed in the electrospray ionization-positive (ESI+) mode, whereas eluents A (water containing 5 mmol/L ammonium formate) and B (acetonitrile) were used in the ESI negative (ESI−) mode. The flow rate was 0.25 mL/min. From the start to 1 minute, D for the ESI+ mode (or B for ESI−) was held at 2%, linearly increased to 50% for the next 9 minutes, linearly increased to 98% for an additional 3 minutes, and then kept constant for 1.5 minutes. Subsequently, D (or B) was returned to 2% in 0.5 minutes and held for an additional 6 minutes before being returned to the initial conditions. The mass spectrometer was run in polarity switching mode (+3.50 kV/−2.5 kV) with an m/z window ranging from 81 to 1,000. The instrument conditions were as follows: capillary temperature, 325 ℃; sheath gas flow, 30 (arbitrary units); auxiliary gas flow, 10 (arbitrary units); full scan resolution, 70,000; and collision voltage, 30 eV. After the instrument calibration, a quality control (QC) sample was run every 5 injections once to ensure the stability of the UPLC-MS system, which was prepared by mixing 10 µL of solution from each sample.
Data preprocessing
The raw data were processed by data transformation and normalization using the ProteoWizard (v3.0.9; Mallick Lab., Stanford, CA, USA) and XCMS (1.42.0; Suizdak Lab., Scripps Research, San Diego, CA, USA) software packages based on the sample retention time, m/z value, and intensity. Multivariate data analyses, such as principal component analysis (PCA), partial least-squared discriminant analysis (PLS-DA), and orthogonal partial least-squared discriminant analysis (OPLS-DA), were performed by SIMCA 14. 1 software (Sartorius Stedim Biotech, Göttingen, Germany). The parameters of the models, such as the R2X, R2Y, and Q2Y, and the R2Y, Q2Y-intercepts, were analyzed to ensure the quality of the multivariate models and to avoid the risk of overfitting. Hierarchical cluster analysis was conducted using the R software package (v3.3.2; the R Foundation for Statistical Computing, Vienna, Austria), and the correlation network was constructed using Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic library and MetaboAnalyst software (Wishart Research Group, Edmonton, Alberta, Canada) (21).
Statistical analysis
Data were presented as mean ± standard deviation. The software SPSS 13.0 (IBM Corp., Chicago, IL, USA) was used for one-way analysis of variance (ANOVA) and Student’s t-test analysis. A P value <0.05 was considered statistically significant.
Results
DHA inhibits SH-SY5Y cell proliferation
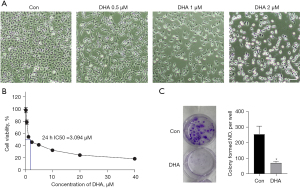
After SH-SY5Y cells were treated with DHA, morphological changes of the cells were observed under a phase contrast microscope. As shown in Figure 1, the cell density after DHA administration was significantly lower than that of the control group. To test the antiproliferative effect of DHA in vitro, SH-SY5Y cells were exposed to DHA (0.5, 1, and 2 µM) for 24 hours. As expected, we observed that the number of cells treated with DHA decreased with the increase in DHA concentration (Figure 1A). The MTT assay was then conducted to assess cell viability. It was shown that DHA significantly inhibited the growth of SH-SY5Y cells, and the inhibition rate also increased with the increase in DHA concentration (Figure 1B). The results showed that the cytotoxicity of DHA in vitro with a half-maximal inhibitory concentration (IC50) of 3 µM. The DHA treatment also completely inhibited the clonal growth of SH-SY5Y (Figure 1C), further supporting its antiproliferation effect. These results showed that DHA inhibited the proliferation of SH-SY5Y in a dose-dependent manner.
DHA induces apoptosis of SH-SY5Y cell
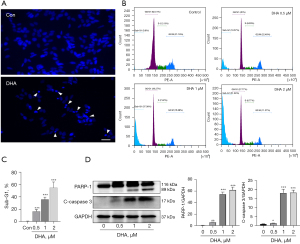
We used DAPI staining to observe the apoptotic nuclear morphology of SH-SY5Y cells. As shown in Figure 2A, DHA treatment caused typical apoptotic nuclear changes, including fragmented nuclei and condensed chromatin (shown as intense blue fluorescence). The DAPI staining showed that 2 µM DHA significantly induced SH-SY5Y cell karyopyknosis. In addition, SH-SY5Y cells were treated with different concentrations (0, 0.5, 1, and 2 µM) of DHA for 24 hours and harvested for flow cytometric analysis. Apoptotic cells (sub-G1) were 18.57%, 37.38%, and 51.84%, respectively, which was significantly higher than that in the control group (0.8%) (Figure 2B,2C; P<0.0001). To further explore the effect of DHA on apoptosis, we used western blot to detect the apoptosis-related proteins, PARP-1 and caspase 3. The data revealed that DHA significantly increased the cleavage of PARP-1 and caspase 3 compared with the control group (Figure 2D). Taken together, these data demonstrated that DHA induced apoptosis in SH-SY5Y cells.
DHA increases ROS generation and DNA double-strand break (DSB)
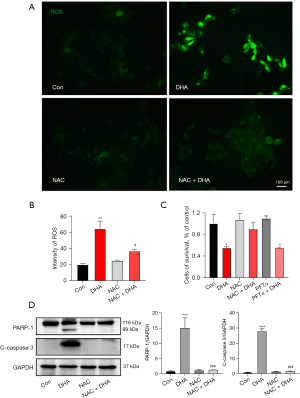
The ROS is an important factor in inducing cell apoptosis. The intracellular ROS in DHA-treated SH-SY5Y cells was determined by DCFH-DA staining. As shown in Figure 3A, DCF fluorescence intensity was significantly increased in SH-SY5Y cells after incubation with 2 µM DHA for 24 hours, indicating that DHA could enhance the intracellular ROS level.

It is known that ROS can damage mitochondria and decrease mitochondrial membrane potential. As shown in Figure 3B, exposure of SH-SY5Y to DHA (2 µM) for 24 hours induced a marked decrease of mitochondrial membrane potential, as detected by JC-1 staining. DSB is one of the most critical DNA lesions related to cell death and genomic integrity. We speculated that DHA could induce DSB to trigger the apoptosis of SH-SY5Y cells. The phosphorylation of H2AX at Ser139, γH2AX, is an early marker in response to DSB. The γH2AX foci on chromosomes represented repaired lesions or unrepaired DNA breaks. Next, we attempted to determine whether DHA can induce γH2AX foci formation in the nucleus. As expected, we observed that DHA treatment could induce the formation of abundant γH2AX foci in SH-SY5Y cells (Figure 3C). In addition, we determined the expression of γH2AX by western blot. The results showed that DHA significantly increased γH2AX expression at 2 µM after 24 hours of treatment (Figure 3D; P<0.01). The results suggested that DHA caused DSB damage via activating γH2AX in SH-SY5Y cells.
Inhibition of ROS can reverse DHA-induced apoptosis
As an ROS inhibitor, NAC was able to resist apoptosis. We used the ROS scavenger NAC to further determine the role of ROS in DHA-mediated apoptosis and the antiproliferation of SH-SY5Y cells. First, ROS generation in SH-SY5Y cells could be inhibited after the treatment with 5 mM NAC (Figure 4A,4B). As shown in Figure 4C, pretreatment with NAC (10 mM) for 3 hours remarkably attenuated DHA-induced apoptosis and growth inhibition in SH-SY5Y cells. Meanwhile, SH-SY5Y cells were treated together with 10 µM PFTα (p53 inhibitor) and 2 µM DHA for 24 hours in the PFTα + DHA group. Interestingly, it was detected that PFTα did not prevent DHA-induced cell death in the PFTα + DHA group (Figure 4C). In addition, the western blot results showed that NAC pretreatment decreased the expression of pro-apoptotic proteins, cleaved caspase 3 and PARP-1 (Figure 4D). The results indicated that DHA-induced apoptosis was not dependent on the p53 pathway in SH-SY5Y cells.
Metabolic profiling
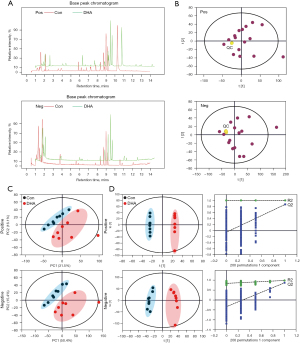
To further study the possible mechanism of DHA inhibiting the proliferation of SH-SY5Y cells, we adopted the strategy of non-targeted cell metabolomics. The typical base peak chromatograms of the control group and DHA group are shown in Figure 5A. Validations of the analysis method, including QC and quality assurance, indicated that this cell metabolomic method was reliable. Furthermore, the PCA score plot exhibited a clear cluster of the pooled QC samples, indicating that the sample analysis sequence had satisfactory stability and repeatability (Figure 5B) (22). Figure 5C shows that the control group and the DHA group are in different regions, indicating that there was a significant metabolic difference between the control and DHA groups. The significant ions from the ESI+ and ESI− modes were merged and imported into the SIMCA-P software package to analyze the metabolic differences between the control and DHA groups. Subsequently, PLS-DA and OPLS-DA were applied to highlight the difference between the control and DHA groups (23). As shown in the OPLS-DA score plot, the control group was clearly separated from the DHA-treatment group (Figure 5D). The permutations plot can help to effectively evaluate whether the OPLS-DA model is overfitting (Figure 5D, right side). When calculating the 2 components; all the blue Q2 points from left to right were lower than the rightmost blue Q2 point. According to the results of the chance permutation, no overfitting was observed. These results excluded the random effects in the constructed model. Using this model, clear separation was produced between the control and DHA groups. These values indicate that there was a significant difference in cell metabolism after drug intervention.
Differential metabolites between the two groups
We used the OPLS-DA model to distinguish the differential metabolites between the control group and the DHA treatment group. A total of 3,206 ions with a variable importance in the project >1 and P<0.05 were selected for subsequent chemical structure identification (24). We used the following steps to identify the chemical structure of the compound. First, the precise molecular weight of the metabolite was identified. Second, the exact mass of the monoisotopic molecular weights was used to search the online databases, such as BioDeepDB (www.biodeep.cn/), the Human Metabolome Database (http://www.hmdb.ca/), the Mass Bank (www.massbank.jp/), Metlin (http://metlin.scripps.edu/), LipidMaps (www.lipidmaps.org/), and mzCloud (www.mzcloud.org/). Subsequently, 125 metabolites were identified. The names of the compounds are presented in Table S1. The level of change for specific biomarkers was indicated by the fold change in abundance. As shown in Table S1, 92 metabolites were significantly increased and 33 metabolites were significantly decreased in the DHA group compared with the control group (Table S1).
The distributions of the intensities in the DHA group (green dots) and the intensities in the control group (red dots) were plotted to discriminate these metabolite alterations (Figure 6). To visualize the relationship between 125 changed metabolites, we drew a metabolite heat map according to the relative change relationship of metabolites (up/down compared with control), and samples were ordered by hierarchical clustering (Figure S1). In general, the changes of these important metabolites in the DHA group were quite different from those in the control group.
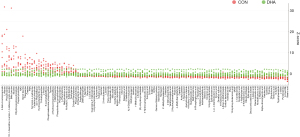
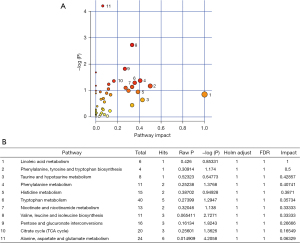
Metabolic pathway analysis in DHA treatment
We further analyzed the functional roles of differential metabolites in cells using the KEGG metabolic library and MetaboAnalyst (25). We evaluated the impact of changes in metabolites on the function of the pathway by changing the critical junction points of the pathway. Using KEGG pathway analysis, 45 metabolic pathways were drawn from the hypergeometric test P-values (vertical axis, shades of red) and impact (horizontal axis, circle diameter) (Figure 7A). Significant changes occurred in the following metabolic pathways: (I) linoleic acid metabolism; (II) phenylalanine, tyrosine, and tryptophan biosynthesis; (III) taurine and hypotaurine metabolism; (IV) phenylalanine metabolism; (V) histidine metabolism; (VI) tryptophan metabolism; (VII) nicotinate and nicotinamide metabolism; (VIII) valine, leucine, and isoleucine biosynthesis; (IX) pentose and glucuronate interconversions; (X) citrate cycle; and (XI) alanine, aspartate and glutamate metabolism (Figure 7B).
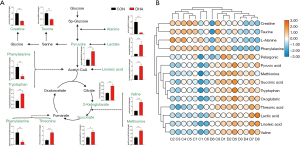
For further analysis, based on the knowledge of these differential metabolites and KEGG database, a network of metabolites associated with the 11 metabolic pathways was constructed (Figure 8A). A heat map of significantly altered metabolites arranged according to direction of change is shown in Figure 8B. The colors (from blue to orange) indicate the relative contents of metabolites in the DHA-treatment group compared with those in the control group.
Discussion
The drug DHA is an active metabolite of sesquiterpene trioxane lactone extracted from Artemisia annua, which is used globally to treat malaria. It has been found to have antiproliferative and pro-apoptotic effects in several tumor cell lines (5,6,26). However, the role of DHA in NB is rarely reported. In the present study, we investigated the antitumor effects of DHA and its possible mechanism in SH-SY5Y cells. Treatment of SH-SY5Y cells with DHA significantly increased the expression of activated caspase 3 and cleaved PARP-1. The cells became rounded and reduced in size following treatment with DHA. Therefore, DHA can induce apoptosis in SH-SY5Y cells. These results suggested that DHA may be a potentially effective drug for the treatment of NB.
The ROS play an important role in cell survival. Low and normal concentrations of ROS are conducive to cell survival, while high concentrations of ROS are harmful to protein, lipid, and DNA (27). ROS-mediated oxidative stress is associated with tumor progression (28). Maintaining ROS within a narrow range allows cancer cells to enhance growth and invasion while limiting their apoptotic susceptibility (29). In the present study, we found that intracellular ROS in SH-SY5Y cells treated with DHA was significantly increased. The ROS inhibitor was able to resist apoptosis. Our results suggested that ROS contributed to SH-SY5Y cell apoptosis induced by DHA.
The H2AX histone protein is rapidly phosphorylated at the serine-139 position (γH2AX) in response to a broad range of DNA lesions (30). Induction of γH2AX is one of the earliest events in the DNA damage response and plays a central role in sensing and repairing DNA damage (31). We found that DHA induced genotoxic stress in SH-SY5Y cells, as indicated by increased γH2AX expression.
The findings of the present study indicated that DHA could induce apoptosis of SH-SY5Y cells. However, there are few reports about the influence of DHA on cell metabolism. Metabolomics analysis can help us further understand the mechanism of action of anti-cancer drugs (32). At present, the antitumor mechanism of DHA at the overall level of cell metabolism has not been reported. The primary goal in the present study was to understand the metabolic features in NB cells after DHA treatment by investigating the metabolic differences using a non-targeted cell metabolomic method. This can be achieved by implementing cellular metabolomic methods to determine the metabolites and metabolic pathways associated with a specific phenotype, and then combining this knowledge with functional and mechanistic biological research (33).
Many nutrients, such as amino acids, are essential for the rapid proliferation of tumor cells. As the most direct raw material for protein synthesis, amino acids can play a variety of roles in energy generation, synthesis of nucleosides, and maintenance of cellular redox homoeostasis (34). In the present study, DHA mainly affected the metabolism of phenylalanine, tryptophan, glycine, serine and threonine, leucine, histidine, and aspartate. The rapid increase of amino acids in SH-SY5Y cells after DHA treatment indicated that the protein synthesis was limited.
The pentose phosphate pathway (PPP), branching from glycolysis, is a major pathway for glucose catabolism (35). The PPP is essential for cell survival and proliferation, and several signaling factors can regulate PPP activity. The precise regulation of this pathway seems to be particularly important for cancer cells. In fact, a previous study has shown that PPP can regulate the reprogramming of energy metabolism in response to oxidation and gene damage (36). Consistent with our findings, the dysregulation of PPP flux has been shown to dramatically impact cancer growth and survival after DHA-treatment (37).
Numerous epidemiological and animal experimental studies have shown that polyunsaturated fatty acids (PUFAs) have a growth-inhibitory effect on mammary (38) and colorectal cancer (39). One of the important PUFAs commonly present in diets is linoleic acid. In vivo and in vitro studies have suggested that linoleic acid induces apoptosis in cancer cells by free radical generation and mitochondrial dysfunction (40,41). Our data showed that linoleic acid was significantly increased after DHA treatment.
In PC12 cells, taurine can protect cells from oxidative stress induced by H2O2 (42). Taurine also reduces ROS by increasing the level of Nrf2 and Trx in HepG2 cells (43). Taurine attenuates mitochondrial oxidative stress (44). Taurine deficiency can result in mitochondrial dysfunction (45). In metabolomic analysis, it was found that DHA treatment reduces the level of taurine in SH-SY5Y cells. A previous report has reported that taurine has antioxidant effects (46). This may account for oxidative stress in SH-SY5Y cells induced by DHA.
As shown by our research results, DHA can inhibit the proliferation and induce apoptosis of SH-SY5Y through energy metabolism, amino acid metabolism, linoleic acid metabolism, and taurine and hypotaurine metabolism, and so on. This lays the foundation for the application of cell metabolomics to interpret the metabolic pathway of NB more accurately at the cellular level.
Conclusions
In the present study, we investigated the inhibitory effect of DHA on the proliferation of SH-SY5Y cells. On the one hand, we found that DHA may restrain cell proliferation and advance cell apoptosis in SH-SY5Y cells, and further explored its potential mechanisms. We reported that DHA induces ROS generation and increases PARP-1 and caspase3 levels in SH-SY5Y cells at cell and molecular levels. On the other hand, using the non-targeted cell metabolomic approaches, our findings demonstrated that DHA can control proliferation and promote apoptosis in SH-SY5Y cells, which were related to 125 proposed metabolites, and linoleic acid metabolism, taurine and hypotaurine metabolism, pentose phosphate metabolism, as well as amino acids metabolism. Metabolomic profiling could identify novel biomarkers of NB. Add functional studies of key metabolites could provide candidate targets for NB therapy. It is helpful to elucidate the mechanism of DHA inhibiting the growth of SHSY5Y cell from the metabolic level. Metabolomics is a valuable tool that can deepen our understanding of the mechanisms of various physiological conditions and abnormal processes, as well as the inhibitory effect of DHA on tumor cell proliferation.
Acknowledgments
Funding: This work was supported by the National Natural Science Foundation of China (No. 82101331), Suzhou Science and Technology Foundation (Nos. SKJY2021087, SYSD2020184, SYS2019058, SS2019042), and the Talent Promotion Project of the Second Affiliated Hospital of Suzhou University (Nos. XKTJ-XK202010, XKTJ9RC202013).
Footnote
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-22-331/rc
Data Sharing Statement: Available at https://tp.amegroups.com/article/view/10.21037/tp-22-331/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-22-331/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Maris JM. Recent advances in neuroblastoma. N Engl J Med 2010;362:2202-11. [Crossref] [PubMed]
- Zafar A, Wang W, Liu G, et al. Targeting the p53-MDM2 pathway for neuroblastoma therapy: Rays of hope. Cancer Lett 2021;496:16-29. [Crossref] [PubMed]
- Zafar A, Wang W, Liu G, et al. Molecular targeting therapies for neuroblastoma: Progress and challenges. Med Res Rev 2021;41:961-1021. [Crossref] [PubMed]
- Efferth T, Oesch F. The immunosuppressive activity of artemisinin-type drugs towards inflammatory and autoimmune diseases. Med Res Rev 2021;41:3023-61. [Crossref] [PubMed]
- Paccez JD, Duncan K, Sekar D, et al. Dihydroartemisinin inhibits prostate cancer via JARID2/miR-7/miR-34a-dependent downregulation of Axl. Oncogenesis 2019;8:14. [Crossref] [PubMed]
- Li S, Huang P, Gan J, et al. Dihydroartemisinin represses esophageal cancer glycolysis by down-regulating pyruvate kinase M2. Eur J Pharmacol 2019;854:232-9. [Crossref] [PubMed]
- Wang L, Li J, Shi X, et al. Antimalarial Dihydroartemisinin triggers autophagy within HeLa cells of human cervical cancer through Bcl-2 phosphorylation at Ser70. Phytomedicine 2019;52:147-56. [Crossref] [PubMed]
- Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov 2009;8:579-91. [Crossref] [PubMed]
- Efferth T. From ancient herb to modern drug: Artemisia annua and artemisinin for cancer therapy. Semin Cancer Biol 2017;46:65-83. [Crossref] [PubMed]
- Dou C, Ding N, Xing J, et al. Dihydroartemisinin attenuates lipopolysaccharide-induced osteoclastogenesis and bone loss via the mitochondria-dependent apoptosis pathway. Cell Death Dis 2016;7:e2162. [Crossref] [PubMed]
- Lin R, Zhang Z, Chen L, et al. Dihydroartemisinin (DHA) induces ferroptosis and causes cell cycle arrest in head and neck carcinoma cells. Cancer Lett 2016;381:165-75. [Crossref] [PubMed]
- Dai X, Zhang X, Chen W, et al. Dihydroartemisinin: A Potential Natural Anticancer Drug. Int J Biol Sci 2021;17:603-22. [Crossref] [PubMed]
- Diepstraten ST, Anderson MA, Czabotar PE, et al. The manipulation of apoptosis for cancer therapy using BH3-mimetic drugs. Nat Rev Cancer 2022;22:45-64. [Crossref] [PubMed]
- Cui Q, Wang JQ, Assaraf YG, et al. Modulating ROS to overcome multidrug resistance in cancer. Drug Resist Updat 2018;41:1-25. [Crossref] [PubMed]
- Gao N, Budhraja A, Cheng S, et al. Interruption of the MEK/ERK signaling cascade promotes dihydroartemisinin-induced apoptosis in vitro and in vivo. Apoptosis 2011;16:511-23. [Crossref] [PubMed]
- Kong R, Jia G, Cheng ZX, et al. Dihydroartemisinin enhances Apo2L/TRAIL-mediated apoptosis in pancreatic cancer cells via ROS-mediated up-regulation of death receptor 5. PLoS One 2012;7:e37222. [Crossref] [PubMed]
- Lu JJ, Chen SM, Ding J, et al. Characterization of dihydroartemisinin-resistant colon carcinoma HCT116/R cell line. Mol Cell Biochem 2012;360:329-37. [Crossref] [PubMed]
- Bauermeister A, Mannochio-Russo H, Costa-Lotufo LV, et al. Mass spectrometry-based metabolomics in microbiome investigations. Nat Rev Microbiol 2022;20:143-60. [Crossref] [PubMed]
- Muthubharathi BC, Gowripriya T, Balamurugan K. Metabolomics: small molecules that matter more. Mol Omics 2021;17:210-29. [Crossref] [PubMed]
- Jang WJ, Choi B, Song SH, et al. Multi-omics analysis reveals that ornithine decarboxylase contributes to erlotinib resistance in pancreatic cancer cells. Oncotarget 2017;8:92727-42. [Crossref] [PubMed]
- Smith CA, Want EJ, O'Maille G, et al. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem 2006;78:779-87. [Crossref] [PubMed]
- Dunn WB, Broadhurst D, Begley P, et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc 2011;6:1060-83. [Crossref] [PubMed]
- Thévenot EA, Roux A, Xu Y, et al. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses. J Proteome Res 2015;14:3322-35. [Crossref] [PubMed]
- Huang Q, Tan Y, Yin P, et al. Metabolic characterization of hepatocellular carcinoma using nontargeted tissue metabolomics. Cancer Res 2013;73:4992-5002. [Crossref] [PubMed]
- Chong J, Xia J. Using MetaboAnalyst 4.0 for Metabolomics Data Analysis, Interpretation, and Integration with Other Omics Data. Methods Mol Biol 2020;2104:337-60. [Crossref] [PubMed]
- Bhaw-Luximon A, Jhurry D. Artemisinin and its derivatives in cancer therapy: status of progress, mechanism of action, and future perspectives. Cancer Chemother Pharmacol 2017;79:451-66. [Crossref] [PubMed]
- Wang Y, Qi H, Liu Y, et al. The double-edged roles of ROS in cancer prevention and therapy. Theranostics 2021;11:4839-57. [Crossref] [PubMed]
- Chang CH, Pauklin S. ROS and TGFβ: from pancreatic tumour growth to metastasis. J Exp Clin Cancer Res 2021;40:152. [Crossref] [PubMed]
- Rodic S, Vincent MD. Reactive oxygen species (ROS) are a key determinant of cancer's metabolic phenotype. Int J Cancer 2018;142:440-8. [Crossref] [PubMed]
- Chen Q, Bian C, Wang X, et al. ADP-ribosylation of histone variant H2AX promotes base excision repair. EMBO J 2021;40:e104542. [Crossref] [PubMed]
- Arnould C, Rocher V, Finoux AL, et al. Loop extrusion as a mechanism for formation of DNA damage repair foci. Nature 2021;590:660-5. [Crossref] [PubMed]
- Schmidt DR, Patel R, Kirsch DG, et al. Metabolomics in cancer research and emerging applications in clinical oncology. CA Cancer J Clin 2021;71:333-58. [Crossref] [PubMed]
- Johnson CH, Ivanisevic J, Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol 2016;17:451-9. [Crossref] [PubMed]
- Vettore L, Westbrook RL, Tennant DA. New aspects of amino acid metabolism in cancer. Br J Cancer 2020;122:150-6. [Crossref] [PubMed]
- Cherepanova OA, Byzova TV. Pentose phosphate pathway drives vascular maturation. Nat Metab 2022;4:15-6. [Crossref] [PubMed]
- Zhang Y, Meng Q, Sun Q, et al. LKB1 deficiency-induced metabolic reprogramming in tumorigenesis and non-neoplastic diseases. Mol Metab 2021;44:101131. [Crossref] [PubMed]
- Leopold JA, Dam A, Maron BA, et al. Aldosterone impairs vascular reactivity by decreasing glucose-6-phosphate dehydrogenase activity. Nat Med 2007;13:189-97. [Crossref] [PubMed]
- Garcia-Hernandez A, Leal-Orta E, Ramirez-Ricardo J, et al. Linoleic acid induces secretion of extracellular vesicles from MDA-MB-231 breast cancer cells that mediate cellular processes involved with angiogenesis in HUVECs. Prostaglandins Other Lipid Mediat 2021;153:106519. [Crossref] [PubMed]
- Ogata R, Mori S, Kishi S, et al. Linoleic Acid Upregulates Microrna-494 to Induce Quiescence in Colorectal Cancer. Int J Mol Sci 2021;23:225. [Crossref] [PubMed]
- Schuster S, Johnson CD, Hennebelle M, et al. Oxidized linoleic acid metabolites induce liver mitochondrial dysfunction, apoptosis, and NLRP3 activation in mice. J Lipid Res 2018;59:1597-609. [Crossref] [PubMed]
- Brown ZJ, Fu Q, Ma C, et al. Carnitine palmitoyltransferase gene upregulation by linoleic acid induces CD4+ T cell apoptosis promoting HCC development. Cell Death Dis 2018;9:620. [Crossref] [PubMed]
- Pan C, Giraldo GS, Prentice H, et al. Taurine protection of PC12 cells against endoplasmic reticulum stress induced by oxidative stress. J Biomed Sci 2010;17:S17. [Crossref] [PubMed]
- Bai J, Yao X, Jiang L, et al. Taurine protects against As2O3-induced autophagy in livers of rat offsprings through PPARγ pathway. Sci Rep 2016;6:27733. [Crossref] [PubMed]
- Castelli V, Paladini A, d'Angelo M, et al. Taurine and oxidative stress in retinal health and disease. CNS Neurosci Ther 2021;27:403-12. [Crossref] [PubMed]
- Bhattacharjee A, Prajapati SK, Krishnamurthy S. Supplementation of taurine improves ionic homeostasis and mitochondrial function in the rats exhibiting post-traumatic stress disorder-like symptoms. Eur J Pharmacol 2021;908:174361. [Crossref] [PubMed]
- Baliou S, Adamaki M, Ioannou P, et al. Protective role of taurine against oxidative stress Mol Med Rep 2021;24:605. (Review). [Crossref] [PubMed]


