Phenotypes and genotypes of the chromosomal instability syndromes
Introduction
Chromosomal instability syndromes (CIS), are also known as chromosomal breakage syndromes are a group of genetic disorders that are typically transmitted in an autosomal recessive pattern of inheritance defined on the basis of cell culture in vitro that the affected individuals exhibit elevated rates of chromosomal breakage or instability, leading to chromosomal rearrangements. CIS often lead to an increased tendency to develop certain types of malignancies. There are several diseases/disorders characterised and classified as chromosomal instability syndromes so far and commonly known as: Fanconi anaemia, Nijmegen breakage syndrome, Bloom syndrome, Ataxia telangiectasia and Ataxia telangiectasia-like disorder (1).
The common cause of chromosomal instability syndromes is the defects of genomic maintenance and DNA repair they are overlapping and sharing some clinical features as summarised in Table 1.

Full table
Studies on CIS for better therapies have achieved exciting successes not only beneficial to CIS self but also beneficial to other diseases. One of the CIS, Fanconi anemia is the disease achieved a success in stem cell transplantation using umbilical blood in 1988 (2) and as an effective therapy for many different types of diseases most commonly in malignancies in clinical application since then.
In addition, studies on CIS found that majority of them are associated with cancers and such studies have explored many mysteries in cancer research. For example, Fanconi anemia is found to associate with many different types of cancers from those 19 responsible mutated genes and 5 of them are associated with breast cancer (3). It is not surprising that studies on Fanconi anemia as an example in CIS is named as the paradigm in the studies in cancer and aging (4).
Fanconi anemia (FA)
FA is the most common and representative type of chromosomal instability syndromes. FA is usually inherited as an autosomal recessive trait, but it can be an X-linked recessive in the subtype B. Patients with FA are characterised with progressive bone marrow failure which it is named as one type of inherited bone marrow failure syndromes, congenital abnormalities and susceptibility to malignancies, commonly in MDS, AML, and solid tumors, commonly in carcinoma of the oropharynx and skin, head and neck squamous cell carcinoma, liver tumors, vaginal squamous cell carcinoma and brain tumors. Individuals with FA also can suffer one, or more types of cancers (5,6). Cells from FA patients exhibit a hypersensitivity to DNA interstrand cross-linking agents such as mitomycin (MMC) and diepoxybutane (DEB). Studies found the association of FA with a pattern of recurrent chromosomal abnormalities including monosomy chromosome 7, deletion of the long arm of chromosome 7, gain of the long arms of chromosome 3 and 1 and the RUNX1 gene mutations in about 20% of the combined MDS cases. Studies showed that chromosome abnormalities with 7 and 3 had a poor prognostic indication value (7).
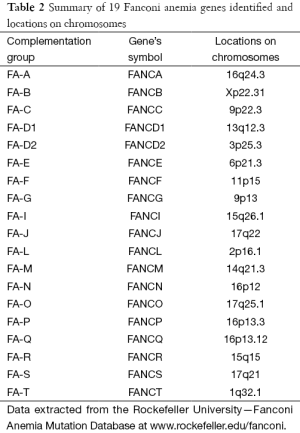
A total of 19 subtypes/complementation groups have been classified and 19 responsible genes have been identified respectively (3) as summarised in Table 2. FA genes encoded proteins participate in the DNA repair. Eight (FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL and FANCM) of the 19 FA proteins formed a FA core complex interacting each other to active the FANCI-FANCD2 protein complex to a monoubiquitinated form to interact with DNA repair proteins (8,9). Five (FANCD1 known as BRCA2 and FANCS known as BRCA1), FANCJ, FANCN and FANCO) of the 19 FA genes are found to associate with breast cancer (10). These findings made a link between FA and cancer through DNA repair mechanism.
Full table
Nijmegen syndrome (NS)
The name of NS derives from the Dutch city Nijmegen where the condition was first described. It also named as Berlin breakage syndrome, Ataxia-Telangiectasia variant 1. It is an autosomal recessive inherited disease with a complex health problematic conditions typically characterised by short stature, microcephaly, distinctive facial feature, recurrent respiratory tract infections, mental development delay from infancy to childhood, dys-functional immune deficiency in T cells and low level of immunoglobulin G and A and increased susceptibility to infections. Individuals with NS increased risk of cancer development (>50 times), commonly in Hodgkin lymphoma, brain tumor, rhabdomyosarcoma about 40% of the affected individuals and usual before age 15. Studies showed heterozygous mutation increase cancer occurrence as well (11).
Most individuals with NS have West Slavic origins and the largest number of them live in Poland.
It is estimated that the prevalence of Nijmegen Syndrome is in approximately 100,000 newborns although the exact data is still unknown (12).
In the clinical presentation and laboratory diagnostic testing, Nijmegen Syndrome and Fanconi anemia show biological overlap. A positive result of chromosomal breakage induced with clastogens such as MMC and DEB can be seen both in Fanconi anemia and Nijmegen Syndrome. Translocations or inversions between chromosomes 7 and 14 can be seen its feature in Nijmegen Syndrome (13).
The genetic cause of NS is due to the mutation of NBN gene mutation with homozygous c.657_661del5 on chromosome 8q21.3, resulted in nibrin protein dys-function which it is involved in several critical cellular functions including the repair of damaged DNA to maintain the stability of the genomic function when breaks of DNA strands are happened in the stage of the genetic material in chromosomes exchanged for cell division. As a result, affected individuals are sensitive to radiation and other agent exposures (14,15). The molecular tests to confirm the diagnosis of a suspected proband are the analysis of exon 6 to determine if the c.657_661 del5 allele and the analysis of entire NBN gene by the sequencing method.
Bloom syndrome (BSyn)
BSyn also named as Bloom-Torre-Machacek syndrome, is a congenital telangiectatic syndrome with the autosomal recessive pattern characterised with short stature, learning disability, a skin rash, sensitive to sun exposure, serious medical complications such as mild immune-deficiency, chronic obstructive pulmonary disease, varying degree of infertile in both male and female, increased risk of diabetes. Increased risk of cancers to 5–8 folds in earlier life, commonly seen myelodysplasia, leukemia, lymphoma, adenocarcinoma and other types of cancers in epithelial tissues is the characteristics of BYyn as well (16). Cytogenetics findings are the aberrant chromosomal rearrangements including quadri-radial, chromatid gaps and breaks, increased frequency of sister chromatid exchange (SCE) from the cultured lymphocytes (17).
Molecular studies demonstrated that mutation of BLM gene which it is a 4,528-bp cDNA sequence defines BLM containing a long open reading frame encoding a 1,417-amino acid protein with 22 exons and is located on chromosome 15q26.1 resulted in RecQ helicase dysfunction in BLM protein is the cause of this disease. The BLM protein helps to maintain genome stability and integrity as the caretakers of the genome and also to prevent the excess sister chromatid exchanges as well (18-21). As a result, SCE is increased to 10 folds under the condition BLM gene mutated. In addition, chromosomal breakage is increased in individuals with Bloom syndrome (22-25).
Ataxia telangiectasia
Ataxia telangiectasia is an autosomal recessive inherited disorder. It was first described in 1926 by two French physicians, Ladislav Syllaba och Kamil Henner (26). It affected 1 in 40,000 to 100,000 people worldwide (27). Ataxia telangiectasia with several complementation groups is also known as Boder-Sedgwick syndrome or Louis–Bar syndrome and its characteristics including a progressive loss of muscular coordination (ataxia), small cerebellum observed by MRI, increased alpha-fetal protein level and dilated blood vessels in the skin (telangiectasia) caused by a defect in ATM gene which it. It also affects the nervous, immune and other body systems (28). The ATM gene provides instructions for making the phosphatidylinositol 3-kinase protein to helps control cell division in the normal development and DNA repair (29-31).
Studies demonstrated that increased cancer risk including T-cell leukemia, B-cell type of lymphoma usually, other types of cancers such as ovarian, breast, gastric cancers, melanoma and sarcoma have been reported (32).
Molecular studies revealed the mutations in the ATM gene with several allelic variants located on chromosome 11q23 are responsible for Ataxia-telangiectasia due to the defects in providing instructions for making the specific protein to help in controlling of cell division, DNA repair and in the normal biological development and function of the body particularly in nervous and immune systems.
Ataxia telangiectasia-like disorder (ATLD)
ATLD is a rare autosomal recessive disorder characterised by progressive cerebellar degeneration that share many clinical presentations with Ataxia telangiectasia but without immune deficiency and telangiectasia, no cancer case report found. It was first designed in 1999 (33) and molecular studies showed that ATLD is caused by inactivating mutations of genes in either homozygous or compound heterozygous (34). ATLD is usually diagnosed at young at the age starting to walk lacking of coordination and imbalance (35).
Studies showed that there are two mutated genes responsible for ATLD either in homozygous or compound heterozygous have been identified. The first is MER11A gene on chromosome 11q21 (ATLD-1) more than 10 different types of variants and the other one is PCNA gene on chromosome 20p12 (33). Cells from patients demonstrated increased susceptibility to radiation due to the defect of DNA repair pathway.
The ATM and MER11A genes are located on the long arm of chromosome 11closely and the biological function of MERA11 protein is linked to Nbs1 in DNA repair.
Acknowledgements
None.
Footnote
Conflicts of Interest: The author has no conflicts of interest to declare.
References
- Taylor AM. Chromosome instability syndromes. Best Pract Res Clin Haematol 2001;14:631-44. [Crossref] [PubMed]
- Gluckman E, Broxmeyer HA, Auerbach AD, et al. Hematopoietic reconstitution in a patient with Fanconi's anemia by means of umbilical-cord blood from an HLA-identical sibling. N Engl J Med 1989;321:1174-8. [Crossref] [PubMed]
- Fanconi Anemia Mutation Database. Available online: www.rockefeller.edu/fanconi
- Schindler D, Hoehn H. Fanconi Anemia: A Paradigmatic Disease for the Understanding of Cancer and Aging (Monographs in Human Genetics, Vol. 15) Basel: S. Karger; 1 edition. 2007.
- Alter BP. Diagnosis, genetics, and management of inherited bone marrow failure syndromes. Hematology Am Soc Hematol Educ Program 2007.29-39. [Crossref] [PubMed]
- Fanconi G. Familiäre infantile perniziosaartige Anämie (pernizioses Blutbild und Konstitution). Jahrb Kinderh 1927;117:257-80.
- Quentin S, Cuccuini W, Ceccaldi R, et al. Myelodysplasia and leukemia of Fanconi anemia are associated with a specific pattern of genomic abnormalities that includes cryptic RUNX1/AML1 lesions. Blood 2011;117:e161-70. [Crossref] [PubMed]
- de Winter JP, Joenje H. The genetic and molecular basis of Fanconi anemia. Mutat Res 2009;668:11-9. [Crossref] [PubMed]
- Sakaguchi H, Nakanishi K, Kojima S. Inherited bone marrow failure syndromes in 2012. Int J Hematol 2013;97:20-9. [Crossref] [PubMed]
- Howlett NG, Taniguchi T, Olson S, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science 2002;297:606-9. [Crossref] [PubMed]
- Weemaes CM, Hustinx TW, Scheres JM, et al. A new chromosomal instability disorder: the Nijmegen breakage syndrome. Acta Paediatr Scand 1981;70:557-64. [Crossref] [PubMed]
- Dembowska-Baginska B, Perek D, Brozyna A, et al. Non-Hodgkin lymphoma (NHL) in children with Nijmegen Breakage syndrome (NBS). Pediatr Blood Cancer 2009;52:186-90. [Crossref] [PubMed]
- Demuth I, Digweed M. The clinical manifestation of a defective response to DNA double-strand breaks as exemplified by Nijmegen breakage syndrome. Oncogene 2007;26:7792-8. [Crossref] [PubMed]
- Lins S, Kim R, Krüger L, et al. Clinical variability and expression of the NBN c.657del5 allele in Nijmegen Breakage Syndrome. Gene 2009;447:12-7. [Crossref] [PubMed]
- Gennery AR, Slatter MA, Bhattacharya A, et al. The clinical and biological overlap between Nijmegen Breakage Syndrome and Fanconi anemia. Clin Immunol 2004;113:214-9. [Crossref] [PubMed]
- Amor-Guéret M. Bloom syndrome, genomic instability and cancer: the SOS-like hypothesis. Cancer Lett 2006;236:1-12. [Crossref] [PubMed]
- Arora H, Chacon AH, Choudhary S, et al. Bloom syndrome. Int J Dermatol 2014;53:798-802. [Crossref] [PubMed]
- Cheok CF, Bachrati CZ, Chan KL, et al. Roles of the Bloom's syndrome helicase in the maintenance of genome stability. Biochem Soc Trans 2005;33:1456-9. [Crossref] [PubMed]
- German J, Sanz MM, Ciocci S, et al. Syndrome-causing mutations of the BLM gene in persons in the Bloom's Syndrome Registry. Hum Mutat 2007;28:743-53. [Crossref] [PubMed]
- Guo RB, Rigolet P, Ren H, et al. Structural and functional analyses of disease-causing missense mutations in Bloom syndrome protein. Nucleic Acids Res 2007;35:6297-310. [Crossref] [PubMed]
- Kaneko H, Kondo N. Clinical features of Bloom syndrome and function of the causative gene, BLM helicase. Expert Rev Mol Diagn 2004;4:393-401. [Crossref] [PubMed]
- Liu Y, West SC. More complexity to the Bloom's syndrome complex. Genes Dev 2008;22:2737-42. [Crossref] [PubMed]
- Ouyang KJ, Woo LL, Ellis NA. Homologous recombination and maintenance of genome integrity: cancer and aging through the prism of human RecQ helicases. Mech Ageing Dev 2008;129:425-40. [Crossref] [PubMed]
- Singh DK, Ahn B, Bohr VA. Roles of RECQ helicases in recombination based DNA repair, genomic stability and aging. Biogerontology 2009;10:235-52. [Crossref] [PubMed]
- Wu L. Role of the BLM helicase in replication fork management. DNA Repair (Amst) 2007;6:936-44. [Crossref] [PubMed]
- Taylor AM, Harnden DG, Arlett CF, et al. Ataxia telangiectasia: a human mutation with abnormal radiation sensitivity. Nature 1975;258:427-9. [Crossref] [PubMed]
- Perlman S, Becker-Catania S, Gatti RA. Ataxia-telangiectasia: diagnosis and treatment. Semin Pediatr Neurol 2003;10:173-82. [Crossref] [PubMed]
- Demuth I, Dutrannoy V, Marques W Jr, et al. New mutations in the ATM gene and clinical data of 25 AT patients. Neurogenetics 2011;12:273-82. [Crossref] [PubMed]
- Verhagen MM, Last JI, Hogervorst FB, et al. Presence of ATM protein and residual kinase activity correlates with the phenotype in ataxia-telangiectasia: a genotype-phenotype study. Hum Mutat 2012;33:561-71. [Crossref] [PubMed]
- McKinnon PJ. ATM and ataxia telangiectasia. EMBO Rep 2004;5:772-6. [Crossref] [PubMed]
- Taylor AM, Byrd PJ. Molecular pathology of ataxia telangiectasia. J Clin Pathol 2005;58:1009-15. [Crossref] [PubMed]
- Hall J. The Ataxia-telangiectasia mutated gene and breast cancer: gene expression profiles and sequence variants. Cancer Lett 2005;227:105-14. [Crossref] [PubMed]
- Stewart GS, Maser RS, Stankovic T, et al. The DNA double-strand break repair gene hMRE11 is mutated in individuals with an ataxia-telangiectasia-like disorder. Cell 1999;99:577-87. [Crossref] [PubMed]
- Delia D, Piane M, Buscemi G, et al. MRE11 mutations and impaired ATM-dependent responses in an Italian family with ataxia-telangiectasia-like disorder. Hum Mol Genet 2004;13:2155-63. [Crossref] [PubMed]
- Hernandez D, McConville CM, Stacey M, et al. A family showing no evidence of linkage between the ataxia telangiectasia gene and chromosome 11q22-23. J Med Genet 1993;30:135-40. [Crossref] [PubMed]


