Genomic diversity in myeloproliferative neoplasms: focus on myelofibrosis
The chronic myeloproliferative neoplasms (MPNs)
The MPNs are a group of clonal haematological disorders that arise from transformation of a multipotent haematopoietic stem cell (1). Chronic myeloid leukaemia (CML) is the best characterised MPN sub category and is characterised by the Philadelphia (Ph) translocation t(Jeny9;22) leading to the BCR/ABL gene fusion associated with abnormal tyrosine kinase activation involved in the causative pathophysiology of CML. The Ph negative, BCR/ABL negative MPNs encompass three distinct clinical sub sets, namely polycythaemia vera (PV), essential thrombocythaemia (ET) and primary myelofibrosis (PMF) that make up the classical MPNs (2). In contrast to CML, disease specific genetic abnormalities have not been detected that distinguish PV, ET and PMF. PMF is the rarest and most complex of the MPNs. Despite this, much advancement in our understanding of the underlying genetic changes in PMF has occurred in recent years that impact patient management.
Classification of primary myelofibrosis (PMF) (Table 1)
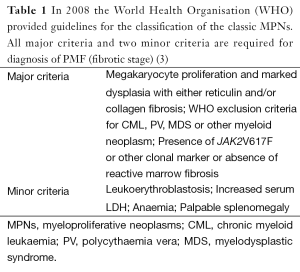
Full table
Disease acceleration is indicated in those patients showing 10-19% blasts, an increased CD34 count with clustering and/or endosteal location on the bone marrow (BM) histology. A blast count above 20% is indicative of transformation to blast phase leukaemia. Patients with PMF may rarely present initially in the accelerated or acute phase (3).
Pre fibrotic/early myelofibrosis (MF)
The World Health Organization (WHO) classification requires, in the absence of reticulin fibrosis, that the megakaryocyte changes must be accompanied by increased marrow cellularity, granulocytic proliferation and decreased erythropoiesis.
Epidemiology of MF
PMF occurs at a frequency of 0.5 to 1.5 per 100,000 (4). It occurs at equal frequency in males and female and the age of onset is usually after the 6th decade. Median reported survival in various studies range between 3.5 to 5 years (4-6). Post MF is found to develop in 25-50% of PV and 2-3% of ET after approximately 15-20 years of follow up (7,8). PMF has been reported rarely in infants and in younger patients with or without a family history of PMF and these cases appear to have a better outcome than adults (4).
Clinical features and diagnosis
Signs and symptoms
Patients usually present with splenomegaly or hepatomegaly as the main physical sign at diagnosis. Profound constitutional symptoms such as fatigue, weight loss, night sweats and fever are common. Thirty percent of patients are asymptomatic at presentation and are detected by an incidental abnormal blood picture or enlarged spleen (9). The disease gradually evolves from the early prefibrotic phase to the fibrotic phase with increasing BM failure.
Laboratory indicators
A large proportion of patients present with anaemia and a haemoglobin level below 100 g/L requiring blood transfusions. Other laboratory findings include leucocytosis or leukopenia, thrombocytosis or thrombocytopenia and circulating myeloblasts, increased serum lactate dehydrogenase and low cholesterol are also observed. Abnormalities on blood film examination at diagnosis may include a leukoerythroblastic blood film, left shifted granulocytes and red cell anisopoikilocytosis. Extra medullary haematopoiesis (EMH) is a striking feature and refers to the presence of proliferating haematopoietic stem or progenitor blood cells outside of the BM (myeloid metaplasia). The most common sites are in the spleen or liver but may arise in the skin, lymph nodes, serosal surfaces, the lungs and spine giving rise to lymphadenopathy, pleural effusion, pneumonia like symptoms or compression of the spinal cord and nerve roots (8,10). A vastly increased number of CD34+ cells are also present in the peripheral blood relative to both normal individuals and to other MPN. An increased number of circulating endothelial progenitors (EPCs) and increased vasculature although not specific to PMF, is a notable feature.
Paediatric PMF
A recent review of PMF in the largest series to date comprising 19 paediatric cases studied over a 30 year period, showed some distinct features compared to that of adult PMF and did not support previous findings that paediatric cases largely resolved spontaneously (11). Paediatric cases albeit rare were infrequently associated with a leukoerythroblastic blood film and 50% of cases showed marked BM eosinophilia not observed in adults. All but one showed one or more cytopaenias and moderate to severe reticulin fibrosis was common together with mild collagen fibrosis in some cases. No osteosclerosis was observed and megakaryocyte dysplasia consisted of separation of nuclear lobes, hypolobation and micromegakaryocytes as opposed to the “cloud like” megakaryocyte nuclei typically observed in adults. No increase in blasts or CD34 count or increased vasculature was observed. All patients required supportive care involving blood or platelet transfusions. Four of nine cases who underwent hematopoietic stem cell transplant (HSCT) died. A further four died of infection. Five cases resolved spontaneously. None transformed to acute phase.
Genetic studies in the above series showed no mutations of the Janus kinase 2 (JAK2 c. 1849G>T) (p.V617F) or myeloproliferative leukaemia viral oncogene [(MPL) c.1544G>T, 1543_1544TG>AA] (p.W515L, p.W515K) usually present in adults. An autosomal recessive inheritance was suspected in some families due to the involvement of siblings and/or consanguinity. The outcome of patients in this series was generally poor and further studies are required to distinguish the pathobiology of spontaneously resolving paediatric cases from those requiring therapy. A subsequent study on calreticulin (CALR) mutations in 14 paediatric cases showed the type 2 CALR c. 1154_1155insTTGTC (p.K385fs*47) mutation to be present in 50% of cases (12) (see section on gene mutations below). These patients showed higher scores for the dynamic international prognostic scoring system plus (DIPSS+) and the international prognostic scoring system (IPSS) models, lower haemoglobin, platelet and total white cell count as well as a lack of splenomegaly and erythroblastosis. No increased eosinophilia was noted and CALR+ cases were all older than 6 years of age. The outcome of the whole cohort was poor with 50% of patients transforming to acute myeloid leukaemia (AML). None had accepted HSCT. The type 1, 52 base pair deletion CALR c.1099_1150del (p.L367fs*46) mutation was not observed in this series and the prognostic significance of CALR mutations in paediatric patients is subject to independent verification.
Prognosis
The median survival in adult PMF is 3.5-5 years but is highly variable and can be estimated using defined prognostic criteria. Younger adult patients with low risk may survive more than a decade while survival is less than 3 months in those that transform to blast phase (13). The current DIPSS+ prognostic model was formulated in 2009 by an international working party (14). The major inclusion in the DIPSS+ model was formal incorporation of cytogenetic information and transfusion dependency that were not previously included to inform risk. In this model, high risk karyotypic results such as complex karyotypes (>3 karyotypic aberrations), or those with sole or 2 of the following: −5/del5q, −7/del7q, +8, inversion 3q, 11q23 abnormalities or del(Jeny12p) as well as transfusion dependency and a platelet count less that 100×109/L were also assigned a score. The DIPSS+ study investigators determined that a proportion of patients in each risk group showed adverse cytogenetics, transfusion dependency or low platelets. Due to the inclusion of these latter criteria, further risk stratification of patients is now possible especially in the intermediate risk groups (15).
Cytogenetics of PMF
Clinical management of PMF is confounded by the extreme variability in disease course. Inter-patient variability also exists with regard to the extent of clonal involvement of the haematopoietic lineages. No monoclonality has however, been demonstrated in the BM fibroblasts (5,16). Clonal chromosome aberrations have been demonstrated in 35-60% of PMF, 15-20% of PV and less than 5% of ET (17,18). The most common changes are del(Jeny13q),del(Jeny20q),+8,+9,+1q and the pattern of abnormalities detected is common to all three MPNs. The complexity of the karyotype tends to increase over time and there is no difference in the type of chromosomal rearrangements observed in PMF and post MF MPN. Multivariate analysis showed +8 and 12p− to be independent poor prognostic markers while 13q− and 20q− showed no adverse effect on survival in multiple studies (19). More recent studies have indicated a monosomal karyotype, −5/del(Jeny5q), −7/del(Jeny7q), 11q23 abnormalities or 3q abnormalities to confer a dismal outcome reflecting the importance of karyotype analysis in clinical management of patients (15).
Single case abnormalities are not uncommon in MPN and the further characterization of these seemingly obscure abnormalities have led to the discovery of highly relevant gene rearrangements such as the ten eleven translocation member 2 (TET2) gene mutation on 4q24, FIP1-like-1/platelet-derived growth factor alpha (FIP1L1/PDGFRA) fusion on 4q12 and also a novel mechanism involving inactivation of the Casitas B-lineage lymphoma (CBL) gene on 11q23 that has proven to be biologically significant in several myeloid diseases with implications for patient therapy (2,20,21). Recent cytogenetics studies reported the association of 1q gain with polyploidy and a trend towards advancing disease in a patient series (22). Patients that transform to acute phase usually show complex karyotypes at transformation and a significantly decreased median survival (23). The role of specific chromosome abnormalities in chronic phase and leukaemic transformation are unknown. Preliminary correlations between the karyotype and specific gene mutations showed a significant association between a normal karyotype and the presence of additional sex combs like 1 (ASXL1) and U2 small nuclear RNA auxiliary factor 1 (U2AF1) mutations (24). A strong correlation was also observed between the del(Jeny20q)/serine-arginine-rich splicing factor 2 (SRSF2) mutation, del(Jeny13q)/CALR mutation and +9/JAK2 mutation.
The use of SNPa added a powerful new dimension to the spectrum of cytogenetic abnormalities that could be detected. The major advantage with SNPa is the ability to provide independent but simultaneous genotype calls and copy number changes. This allows the detection of somatic mosaicism and copy neutral loss of heterozygosity (CNLOH) in a single analysis. The majority of CNLOH involved the 9p arm containing the JAK2 gene, however, other regions were also recurrently altered such as 1p, 4q22, 11q, 14q, 15q and 17q (22,25-27). Non random copy number changes affected the 6p, 8p, 17q11.2 and 22q regions. The detection of copy number abnormalities known to be associated with MPN such as Neurofibromatosis 1 (NF1), 20q and 13q deletions among others were significantly enhanced by SNPa. Novel copy number changes described at leukaemic transformation involved 7q, 16q, 19p, and 21q (28). Failure to detect balanced rearrangements, polyploidy or minor sub clones were the major limitations encountered with SNPa in the absence of a karyotype.
Gene mutations
In 2005 four different groups detected an identical gain of function point mutation of the JAK2 gene (29-32). This mutation causes a valine to phenylalanine substitution (V617F) in exon 14 of the JAK2 gene located on chromosome 9p24. The JAK2V617F gene mutation results in the continuous activation of the JAK-STAT signalling pathway and constitutive expression of STAT3/5 resulting in unchecked myeloproliferation. The incidence of the JAK2 mutation in the classic MPNs ranges between 35-50% in PMF, 57% in ET and 74-97% in PV but the mutation by itself does not explain the distinctly different phenotypes associated with the three disease sub entities (33).
A second mutation in JAK2 exon 12 was discovered exclusively in patients with PV (3%) who are negative for the JAK2V617F mutation. The exon 12 mutation does not alter the typical PV disease phenotype. A number of new mutations in MPN have been described in recent years that are thought to be co-operating mutations needed for disease initiation and progression (34). Table 2 indicates the frequency of these gene mutations in the different disease sub types (33,34,52,53).

Full table
Activating mutations have been ascribed to mutations in JAK2 and the thrombopoietin receptor MPL. The MPL mutation is more specific to PMF and ET and has not been reported in PV (Table 2). The level of the JAK2 mutant allele burden has been shown to be a factor implicated in the MPN sub types. Low levels of JAK2 mutation in PMF are characterised by BM failure, low cell counts, infections and show an inferior outcome compared to those PMF patients with high mutant load or wild type JAK2 that show a more classic myeloproliferative phenotype (54-56). This possible biological sub group bears some significance, as these patients show marked cytopaenias and are likely to be excluded from clinical trials and thus make conclusions about generalised outcomes difficult (57). The above studies underscore the importance of serial studies on JAK2 in these patients in order to elucidate the presence, timing of mutation acquisition and the role of JAK2 allele burden in PMF. Other genes involved with activation of the JAK-STAT pathway include inactivating mutations in the SH2B adaptor protein 3 (SH2B3)/LNK and NF1 genes while dominant negative mutations occur in CBL.
Recently, mutations in the CALR gene have been described in 67% of cases with ET and 86% of cases with PMF that are negative for JAK2 or MPL mutations (48,49). CALR mutations were found to be mutually exclusive of both JAK2 and MPL mutations. The two most common mutations found comprised a 52 base pair deletion (type 1) or the 5 base pair insertion (type 2) in exon 9 of CALR. All mutations resulted in the same one base pair frame shift with the production of an abnormal protein with a novel C terminus. The CALR mutation also showed cytokine independent growth of cells due to activation of signal transducer and activator of transcription 5 (STAT5) involved with the JAK/STAT pathway but its exact role in MPN remains to be clarified. Patients with the CALR mutation showed an improved survival and lower risk of thrombosis compared to patients carrying the JAK2 mutation. Triple negative (JAK2−, MPL−, CALR−) patients make up approximately one third of PMF cases and show an inferior outcome compared to CALR+ cases.
Mutations in splice factor genes U2AF1, SRSF2 and splicing factor 3b, subunit 1 (SF3B1) are a common feature in PMF and occur during the early stages of the disease (58). These are rarely described in association with CALR mutations but are frequently associated with mutations of JAK2 or MPL. U2AF1 and SRSF2 are strongly associated with anaemia and thrombocytopaenia.
Inactivating mutations have been detected in genes involved with transcriptional regulation through methylation repression/de repression of genes. These include ASXL1, enhancer of zeste homolog 2 (EZH2), DNA (cytosine-5) methyltransferase 3 alpha (DNMT3A) and TET2. It has been suggested that these mutations may predate the acquisition of the JAK2 mutation. Low levels of JAK2 mutation in cases with a high disease load determined by other markers of clonality such as X inactivation or cytogenetic abnormalities is further supporting evidence of a pre JAK2 mutation clone (34). Alternatively, the JAK2 mutation may be acquired prior to these epigenetic mutations or may occur concurrently with one or several mutations in PMF as opposed to PV or ET that usually show only one mutation.
In studies of patients with JAK2 positive MPN half the cases that transformed to blast phase did not retain the JAK2 mutation after transformation (59). A possible explanation is the presence of two different haematopoietic stem cell clones that subsequently acquired a JAK2 mutation in one clone and an AML inducing mutation in the other (60,61).
Several mutations have been associated with blast phase MPN that are rare in chronic phase (28). This would provide indicators for an increased risk of disease transformation that may necessitate more aggressive therapy or provide the option of BM transplantation in suitable candidates. Genes in blast phase MPN involve Runt-related transcription factor 1 (RUNX1), tumour protein 53 (TP53), IKAROS zinc finger protein 1 (IKZF1), CBL, Wilms tumour 1 (WT1), Kirsten rat sarcoma viral oncogene (KRAS) and NF1. In addition, isocitrate dehrogenase 1/2 (IDH1/2) are also implicated in disease progression. SRSF2 mutations appear to be most strongly associated with post MPN AML and show a reduced overall survival. The highest risk mutation profile associated with PMF is CALR negative and ASXL1 positive. The inclusion of the gene mutation profile in the DIPSS+ prognostic model is thus becoming increasingly important to identify risk factors for disease transformation and survival in PMF.
Genetic predisposition to MPN
Several lines of evidence have been put forward to support a hypothesis for the genetic predisposition to the development of MPN. This is validated by the markedly increased risk for MPN in first degree relatives of MPN patients that occurs in 5-10% of MPN as well as the bi clonal nature of the disease in some cases (43). Polymorphic SNP variants have been described in the JAK2 gene itself. The 46/1 variant (rs1327494) results in a 3 fold increase in the risk of developing JAK2 positive MPN and a 1.4 fold risk for MPL mutations. The SNP rs2853677 A/G transition in the telomerase reverse transcriptase (TERT) gene is also a strong contender for disease predisposition to MPN.
Patient therapy
Haematopoietic stem cell transplantation is the only curative therapeutic option currently available for PMF. However, this is a high risk procedure and the mortality associated with HSCT is extremely high in patients above 70 years of age making this an unsuitable option for many patients with PMF (62). The majority of therapeutic regimes in clinical practice have been largely empirically derived and are considered mostly palliative (63).
Since the discovery of the JAK2 mutation, emphasis has since shifted to the therapeutic targeting of the abnormal clone (64). Several JAK inhibitors have undergone clinical trials. Of theses the COMFORT (controlled MF study with oral JAK2 treatment) 1 and 11 trials using ruxolitinib has shown superior results in patients with intermediate and high risk MF (65). Participants showed a major relief of constitutional symptoms, a marked decrease (35%) in spleen size and an increase in overall survival. Ruxolitinib does not eradicate the malignant clone but provides a superior palliative care option resulting in an increase in quality of life. The main side effects associated with ruxolitinib include anaemia, thrombocytopaenia and neutropaenia. Selection criteria for treatment may thus exclude some patient subsets such as those with significant BM failure or transfusion dependency. In addition, adverse events such as myelosupression, failure to respond and disease transformation have also been described (66). In addition, availability of the drug to patients on a worldwide scale may be limited due to cost. As an alternative approach, the recent application of gene editing using the CRISPR/cas9 system to precisely target the JAK2V617F mutation in induced pluripotent stem cells, demonstrated a strategy to replace the mutant JAK2 with the normal wild type copy by using the double stranded DNA repair mechanism of the cell (67,68). This represents a major step forward in the quest for more effective therapy in PMF with relevant known mutations and highlights the need for comprehensive genetic screening of patients as well as continued research on the causative molecular defects in MPN.
Acknowledgements
We thank Dr William Stevenson and Professor Chris Ward for their detailed review of the manuscript.
Footnote
Conflicts of Interest: The author has no conflicts of interest to declare.
References
- Skoda R. Advances in the understanding and management of myeloproliferative disorders. Eur J Haematol Suppl 2007;68:2-4. [PubMed]
- Cools J, DeAngelo DJ, Gotlib J, et al. A tyrosine kinase created by fusion of the PDGFRA and FIP1L1 genes as a therapeutic target of imatinib in idiopathic hypereosinophilic syndrome. N Engl J Med 2003;348:1201-14. [PubMed]
- Swerdlow SH. eds. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. Lyon: International Agency for Research on Cancer, 2008.
- Tefferi A. Myelofibrosis with myeloid metaplasia. N Engl J Med 2000;342:1255-65. [PubMed]
- Tefferi A. Pathogenesis of myelofibrosis with myeloid metaplasia. J Clin Oncol 2005;23:8520-30. [PubMed]
- Thiele J, Kvasnicka HM. Myelofibrosis in chronic myeloproliferative disorders--dynamics and clinical impact. Histol Histopathol 2006;21:1367-78. [PubMed]
- Barosi G. Myelofibrosis with myeloid metaplasia: diagnostic definition and prognostic classification for clinical studies and treatment guidelines. J Clin Oncol 1999;17:2954-70. [PubMed]
- Caramazza D, Hussein K, Siragusa S, et al. Chromosome 1 abnormalities in myeloid malignancies: a literature survey and karyotype-phenotype associations. Eur J Haematol 2010;84:191-200. [PubMed]
- Cervantes F, Barosi G. Myelofibrosis with myeloid metaplasia: diagnosis, prognostic factors, and staging. Semin Oncol 2005;32:395-402. [PubMed]
- Miyata T, Masuzawa M, Katsuoka K, et al. Cutaneous extramedullary hematopoiesis in a patient with idiopathic myelofibrosis. J Dermatol 2008;35:456-61. [PubMed]
- DeLario MR, Sheehan AM, Ataya R, et al. Clinical, histopathologic, and genetic features of pediatric primary myelofibrosis--an entity different from adults. Am J Hematol 2012;87:461-4. [PubMed]
- An W, Wan Y, Guo Y, et al. CALR mutation screening in pediatric primary myelofibrosis. Pediatr Blood Cancer 2014;61:2256-62. [PubMed]
- Mesa RA. The dawn of targeted therapy for primary myelofibrosis: opportunities and challenges. Leuk Res 2007;31:883-6. [PubMed]
- Gangat N, Caramazza D, Vaidya R, et al. DIPSS plus: a refined Dynamic International Prognostic Scoring System for primary myelofibrosis that incorporates prognostic information from karyotype, platelet count, and transfusion status. J Clin Oncol 2011;29:392-7. [PubMed]
- Vaidya R, Caramazza D, Begna KH, et al. Monosomal karyotype in primary myelofibrosis is detrimental to both overall and leukemia-free survival. Blood 2011;117:5612-5. [PubMed]
- Vainchenker W, Constantinescu SN. A unique activating mutation in JAK2 (V617F) is at the origin of polycythemia vera and allows a new classification of myeloproliferative diseases. Hematology Am Soc Hematol Educ Program 2005.195-200. [PubMed]
- Gangat N, Strand J, Lasho TL, et al. Cytogenetic studies at diagnosis in polycythemia vera: clinical and JAK2V617F allele burden correlates. Eur J Haematol 2008;80:197-200. [PubMed]
- Bacher U, Haferlach T, Kern W, et al. Conventional cytogenetics of myeloproliferative diseases other than CML contribute valid information. Ann Hematol 2005;84:250-7. [PubMed]
- Tefferi A, Mesa RA, Schroeder G, et al. Cytogenetic findings and their clinical relevance in myelofibrosis with myeloid metaplasia. Br J Haematol 2001;113:763-71. [PubMed]
- Grand FH, Hidalgo-Curtis CE, Ernst T, et al. Frequent CBL mutations associated with 11q acquired uniparental disomy in myeloproliferative neoplasms. Blood 2009;113:6182-92. [PubMed]
- Delhommeau F, Dupont S, Della Valle V, et al. Mutation in TET2 in myeloid cancers. N Engl J Med 2009;360:2289-301. [PubMed]
- Singh NR, Morris CM, Koleth M, et al. Polyploidy in myelofibrosis: analysis by cytogenetic and SNP array indicates association with advancing disease. Mol Cytogenet 2013;6:59. [PubMed]
- Mesa RA, Li CY, Ketterling RP, et al. Leukemic transformation in myelofibrosis with myeloid metaplasia: a single-institution experience with 91 cases. Blood 2005;105:973-7. [PubMed]
- Wassie E, Finke C, Gangat N, et al. A compendium of cytogenetic abnormalities in myelofibrosis: molecular and phenotypic correlates in 826 patients. Br J Haematol 2015;169:71-6. [PubMed]
- Kawamata N, Ogawa S, Yamamoto G, et al. Genetic profiling of myeloproliferative disorders by single-nucleotide polymorphism oligonucleotide microarray. Exp Hematol 2008;36:1471-9. [PubMed]
- Gondek LP, Dunbar AJ, Szpurka H, et al. SNP array karyotyping allows for the detection of uniparental disomy and cryptic chromosomal abnormalities in MDS/MPD-U and MPD. PLoS One 2007;2:e1225. [PubMed]
- Stegelmann F, Bullinger L, Griesshammer M, et al. High-resolution single-nucleotide polymorphism array-profiling in myeloproliferative neoplasms identifies novel genomic aberrations. Haematologica 2010;95:666-9. [PubMed]
- Zhang SJ, Rampal R, Manshouri T, et al. Genetic analysis of patients with leukemic transformation of myeloproliferative neoplasms shows recurrent SRSF2 mutations that are associated with adverse outcome. Blood 2012;119:4480-5. [PubMed]
- Kralovics R, Passamonti F, Buser AS, et al. A gain-of-function mutation of JAK2 in myeloproliferative disorders. N Engl J Med 2005;352:1779-90. [PubMed]
- Baxter EJ, Scott LM, Campbell PJ, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 2005;365:1054-61. [PubMed]
- Jones AV, Kreil S, Zoi K, et al. Widespread occurrence of the JAK2 V617F mutation in chronic myeloproliferative disorders. Blood 2005;106:2162-8. [PubMed]
- Levine RL, Wadleigh M, Cools J, et al. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell 2005;7:387-97. [PubMed]
- Tefferi A. Novel mutations and their functional and clinical relevance in myeloproliferative neoplasms: JAK2, MPL, TET2, ASXL1, CBL, IDH and IKZF1. Leukemia 2010;24:1128-38. [PubMed]
- Vainchenker W, Delhommeau F, Constantinescu SN, et al. New mutations and pathogenesis of myeloproliferative neoplasms. Blood 2011;118:1723-35. [PubMed]
- James C, Ugo V, Le Couédic JP, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 2005;434:1144-8. [PubMed]
- Pikman Y, Lee BH, Mercher T, et al. MPLW515L is a novel somatic activating mutation in myelofibrosis with myeloid metaplasia. PLoS Med 2006;3:e270. [PubMed]
- Beer PA, Campbell PJ, Scott LM, et al. MPL mutations in myeloproliferative disorders: analysis of the PT-1 cohort. Blood 2008;112:141-9. [PubMed]
- Chaligné R, Tonetti C. New mutations of MPL in primitive myelofibrosis: only the MPL W515 mutations promote a G1/S-phase transition. Leukemia 2008;22:1557-66. [PubMed]
- Pardanani A, Lasho T, Finke C, et al. LNK mutation studies in blast-phase myeloproliferative neoplasms, and in chronic-phase disease with TET2, IDH, JAK2 or MPL mutations. Leukemia 2010;24:1713-8. [PubMed]
- Makishima H, Jankowska AM, Tiu RV, et al. Novel homo- and hemizygous mutations in EZH2 in myeloid malignancies. Leukemia 2010;24:1799-804. [PubMed]
- Nikoloski G, Langemeijer SM, Kuiper RP, et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat Genet 2010;42:665-7. [PubMed]
- Ding Y, Harada Y, Imagawa J, et al. AML1/RUNX1 point mutation possibly promotes leukemic transformation in myeloproliferative neoplasms. Blood 2009;114:5201-5. [PubMed]
- Harutyunyan AS, Kralovics R. Role of germline genetic factors in MPN pathogenesis. Hematol Oncol Clin North Am 2012;26:1037-51. [PubMed]
- Pardanani A, Lasho TL, Finke CM, et al. IDH1 and IDH2 mutation analysis in chronic- and blast-phase myeloproliferative neoplasms. Leukemia 2010;24:1146-51. [PubMed]
- Abdel-Wahab O, Pardanani A, Rampal R, et al. DNMT3A mutational analysis in primary myelofibrosis, chronic myelomonocytic leukemia and advanced phases of myeloproliferative neoplasms. Leukemia 2011;25:1219-20. [PubMed]
- Lasho TL, Christy M, Patnaik M, et al. SRSF2 mutations in primary myelofibrosis: significant clustering with IDH mutations and independent association with inferior overall and leukemia free survival. Blood 2012;120:4168-71. [PubMed]
- Jutzi JS, Bogeska R, Nikoloski G, et al. MPN patients harbor recurrent truncating mutations in transcription factor NF-E2. J Exp Med 2013;210:1003-19. [PubMed]
- Klampfl T, Gisslinger H, Harutyunyan AS, et al. Somatic mutations of calreticulin in myeloproliferative neoplasms. N Engl J Med 2013;369:2379-90. [PubMed]
- Nangalia J, Massie CE, Baxter EJ, et al. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N Engl J Med 2013;369:2391-405. [PubMed]
- Bartels S, Lehmann U, Busche G, et al. SRSF2 and U2AF1 mutations in primary myelofibrosis are associated with JAK2 and MPL but not calreticulin mutation and may independently reoccur after allogeneic stem cell transplantation. Leukemia 2015;29:253-5. [PubMed]
- Tefferi A, Finke CM, Lasho TL, et al. U2AF1 mutations in primary myelofibrosis are strongly associated with anemia and thrombocytopenia despite clustering with JAK2V617F and normal karyotype Leukemia 2014;28:431-3. [PubMed]
- Passamonti F, Maffioli M, Caramazza D, et al. Myeloproliferative neoplasms: from JAK2 mutations discovery to JAK2 inhibitor therapies. Oncotarget 2011;2:485-90. [PubMed]
- Campregher PV, Santos FP, Perini GF, et al. Molecular biology of Philadelphia-negative myeloproliferative neoplasms. Rev Bras Hematol Hemoter 2012;34:150-5. [PubMed]
- Guglielmelli P, Barosi G, Specchia G, et al. Identification of patients with poorer survival in primary myelofibrosis based on the burden of JAK2V617F mutated allele. Blood 2009;114:1477-83. [PubMed]
- Hubbeling HG, Frank DM, Hexner EO. Myelofibrosis 2012: it's complicated. Ther Adv Hematol 2012;3:131-46. [PubMed]
- Tefferi A, Lasho TL, Huang J, et al. Low JAK2V617F allele burden in primary myelofibrosis, compared to either a higher allele burden or unmutated status, is associated with inferior overall and leukemia-free survival. Leukemia 2008;22:756-61. [PubMed]
- Verstovsek S, Kantarjian H, Mesa RA, et al. Safety and efficacy of INCB018424, a JAK1 and JAK2 inhibitor, in myelofibrosis. N Engl J Med 2010;363:1117-27. [PubMed]
- Tefferi A, Finke CM, Lasho TL, et al. U2AF1 mutations in primary myelofibrosis are strongly associated with anemia and thrombocytopenia despite clustering with JAK2V617F and normal karyotype. Leukemia 2014;28:431-3. [PubMed]
- Wu YY, Hung HM, Chen TS, et al. Decreased JAK2 V617F allele burden level in a myelofibrosis with myeloid metaplasia patient with leukemic transformation. Leuk Res 2008;32:1783-6. [PubMed]
- Vannucchi AM, Antonioli E, Guglielmelli P, et al. Clinical correlates of JAK2V617F presence or allele burden in myeloproliferative neoplasms: a critical reappraisal. Leukemia 2008;22:1299-307. [PubMed]
- Beer PA, Delhommeau F, LeCouédic JP, et al. Two routes to leukemic transformation after a JAK2 mutation-positive myeloproliferative neoplasm. Blood 2010;115:2891-900. [PubMed]
- Kröger N, Zabelina T, Schieder H. Pilot study of reduced-intensity conditioning followed by allogeneic stem cell transplantation from related and unrelated donors in patients with myelofibrosis. Br J Haematol 2005;128:690-7. [PubMed]
- Cervantes F, Alvarez-Larran A, Hernandez-Boluda J, et al. Erythropoietin treatment of the anaemia of myelofibrosis with myeloid metaplasia: results in 20 patients and review of the literature. British Journal of Haematology 2004;127:399-403. [PubMed]
- Pardanani A, Hood J, Lasho T. TG101209, a selective JAK2 kinase inhibitor, suppresses endogenous and cytokine-supported colony formation from hematopoietic progenitors carrying JAK2V617F or MPLW515K/L mutations. ASH Annual Meeting Abstracts 2006;108:2680.
- Mascarenhas JO, Orazi A, Bhalla KN, et al. Advances in myelofibrosis: a clinical case approach. Haematologica 2013;98:1499-509. [PubMed]
- Tefferi A. JAK inhibitors for myeloproliferative neoplasms: clarifying facts from myths. Blood 2012;119:2721-30. [PubMed]
- Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014;346:1258096. [PubMed]
- Smith C, Abalde-Atristain L, He C, et al. Efficient and Allele-Specific Genome Editing of Disease Loci in Human iPSCs. Mol Ther 2015;23:570-7. [PubMed]


