
Phenotypes and epigenetic errors in patients with Beckwith-Wiedemann syndrome in China
Introduction
Beckwith-Wiedemann syndrome (BWS, OMIM 130650), which is primarily caused by epigenetic errors, has an estimated incidence of 1:26,000 to 1:10,340 live births (1,2). The main clinical features of BWS include macroglossia, hemihypertrophy, abdominal wall defects, multifocal Wilms’ tumor, and prolonged hyperinsulinism; however, considerable phenotypic variability has also been recognized in BWS (3). The epigenetic etiology of BWS is associated with the 11p15.5 region, which harbors a cluster of genes (3). Gene expression in the 11p15.5 region is regulated by two differentially methylated regions: imprinting control (IC) regions IC1 and IC2. Abnormality in IC1 methylation in the maternal allele results in IGF2 overexpression and H19 underexpression (4), whereas decreased maternal IC2 methylation leads to the release of the transcription of non-coding RNA suppression gene KCNQ1OT1, which arrests the expression of the growth-inhibiting genes CDKN1C, KCNQ1, and PHLDA2 (5). Also, paternal uniparental isodisomy (pUPD) in 11p15.5, which combines the above epigenetic errors, can lead to both IGF2 overexpression and decreased CDKN1C expression (6). Loss of methylation (LOM) at IC2 (IC2-LOM) has been reported to cause over 50% of BWS cases, while gain of methylation (GOM) at IC1 (IC1-GOM) occurs in 5–10% of patients. Further, pUPD contributes to the etiology of ~20% of BWS cases. The remaining proportion of BWS cases is attributable to genetic defects, such as mutation of the CDKN1C gene and chromosomal abnormalities, or currently known (epi)genetic anomalies (7).
Patients with BWS have a higher risk of developing life-threatening complications, such as macroglossia, hypoglycemia, and cancer (8-12). Early diagnosis can facilitate the establishment of surveillance protocols, therefore improving the well-being of patients with BWS (13,14). Recent studies have identified a significant difference in the frequencies of clinical characteristics between populations (15); however, other studies have shown high overlap between BWS subtypes (16). Although the ability of recently proposed clinical diagnostic scoring systems to effectively predict positive BWS molecular test results in various populations has been proved (17), questions still surround the efficacy of diagnosing BWS and its subtypes based on patients’ symptoms.
To date, studies on the clinical presentation of BWS have primarily been conducted in Europeans (12,14,16,18-24), while studies in Asian populations are sparse (17,25-28). Therefore, this study aimed to evaluate the efficacy of diagnosing patients with BWS according to their clinical characteristics in a prospective cohort established by symptom screening.
We present the following article in accordance with the STROBE reporting checklist (available at http://dx.doi.org/10.21037/tp-20-243).
Methods
Patients
Between January 2014 and December 2019, a cohort of patients with BWS was recruited according to the following diagnostic criteria: (I) presenting with at least one of the following: macroglossia, hemihypertrophy, or omphalocele; or (II) presenting with at least two of the following: abdominal wall defects (diastasis recti or umbilical hernia), ear creases/pits, facial nevus flammeus (diameter >1 cm), birth weight >90th percentile, neonatal hypoglycemia, or ultrasound-confirmed visceromegaly. The above criteria were formulated to take into account all previously described diagnostic criteria based on the clinical features of BWS [i.e., Elliott et al. 1994 (21), DeBaun et al. 1998 (22), Gaston et al. 2001 (23), Weksberg et al. 2001 (24), and Zarate et al. 2009 (14)].
After the patient was clinically diagnosed with BWS according to the above criteria, additional information was collected, including age, gender, birth history (date, gestational age, birth weight), and age at diagnosis. The patient was then examined again for the full spectrum of BWS symptoms described in the aforementioned diagnostic criteria.
The clinically diagnosed patients were then subjected to epigenetic analysis using methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA, SALSA MS-MLPA probe mix ME030, MRC Holland) for the detection of aberrant imprinting patterns of the two IC regions in 11p15.5 (paternally methylated H19/IGF2 and maternally methylated KCNQ1OT1), as well as changes in genetic dosage (deletions/duplications). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was acquired by parents of all the patients before enrollment. This study was approved by the Medical Ethics Committee of the Children’s Hospital of Fudan University.
Statistical analysis
Statistical analysis was performed using R (version 3.5.0, www.r-project.org). The frequencies of clinical features in BWS patients with positive epigenetic tests in our cohort were recorded. The same information was recorded from previous cohorts from the following countries: Netherlands (11), France (12), Italy (2,16), United Kingdom (18), and US (29). The Asian cohort comprised patients in our cohort and previous Chinese cohorts (17,27,28). The pooled frequency of each clinical feature was estimated using proportion data from the cohorts using a random-effects model with the DerSimonian and Laird estimator for between-study variance (30). Differences between the European and Asian populations were compared using a fixed-effects model with pooled proportion estimates generated by the above random-effects model and separate between-study variance within each population.
The diagnostic efficacy of clinical characteristics was evaluated in our cohort using receiver operating characteristic (ROC) curve analysis for the following historical criteria: (I) Elliott et al. 1994 (21), (II) Debaun et al. 1998 (22), (III) Gaston et al. 2001 (23), (IV) Weksberg et al. 2001 (24), and (V) Zarate et al. 2009 (14). The diagnostic efficacy of two recently developed scoring systems, Ibrahim et al. 2014 (18) and Brioude et al. 2018 (3), was also assessed. Furthermore, multivariate prediction model analysis with epigenetic diagnosis (a positive result of a methylation assay test against all clinically diagnosed patients, or a subtype of epigenetic error against a positive result of a methylation assay test) as a dependent variable was conducted. Logistic regression analysis was performed to assess the association between each clinical feature and epigenetic diagnosis. ROC curves were also plotted to compare the diagnostic efficacy of each clinical feature to that of epigenetic diagnosis. The regression coefficients and area under the ROC curve (AUC) of each clinical feature were used to develop a weighted scoring system to determine the best AUC for the sum of the scores in the prediction of epigenetic diagnosis in our BWS cohort.
Results
During this 6-year study, 97 patients who fulfilled our diagnostic criteria for BWS based on clinical features were identified. The birth weight of the patients in our cohort was 3.4±0.8 (range, 1.7–5.3) kg, and 51 (52.6%) were female. Among these patients, 66 (68%) were confirmed to have epigenetic errors. The mean age at clinical diagnosis and genetic diagnosis was 4.83 (range, 0.03–60) months and 5.38 (range, 0.3–60.6) months, respectively. The patients received their epigenetic test results after 0.78 (range, 0.23–2) months (Table 1).
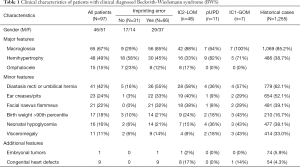
Full table
Figure 1 shows the estimated frequencies of the epigenetic subtypes and clinical features of BWS patients in European and Asian populations. The most common epigenetic subtype among BWS patients was IC2-LOM [66.0%, 95% confidence interval (CI): 61.8–70.1%], followed by pUPD (23.6%, 95% CI: 19.5–27.6%) and IC1-GOM (9.8%, 95% CI: 8.3–11.2%). No significant differences were observed in the distribution of epigenetic subtypes between European and Asian populations. The most common clinical features reported in BWS patients were macroglossia (83.2%, 95% CI: 77.8–88.6%), abdominal wall defects (71.3%, 95% CI: 61.7–81.0%), and ear creases/pits (55.3%, 95% CI: 47.5–63.0%). There was substantial heterogeneity (I2>50%) between the studies in most estimations. No differences were found in the frequencies of major clinical features between European and Asian populations, apart from the occurrence of neonatal hypoglycemia being significantly higher in European patients [47.8% (95% CI: 36.9–58.7%) vs. 25.9% (95% CI: 15.1–36.6%); P=0.002].

The frequency of clinical features differed between epigenetic subtypes of BWS (Figure 2). Patients with IC2-LOM had significantly higher frequencies of ear creases/pits and facial nevus flammeus than the other two epigenotypes. Patients with pUPD showed significantly less macroglossia but had more hemihypertrophy. Visceromegaly occurred significantly more frequently in patients with the IC1-GOM subtype. Further analysis among the epigenetic subtypes of BWS did not reveal a significant difference in the distribution of clinical features between European and Asian populations (data not shown).
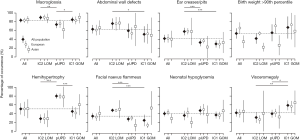
Scoring system and diagnostic efficacy using clinical features in BWS
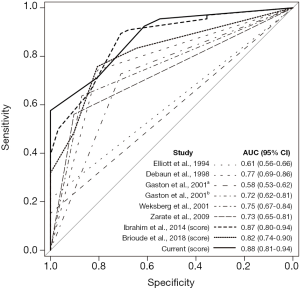
The AUC was comparably good for the recently developed scoring systems [0.87 for Ibrahim et al., 2014 (18) and 0.82 for Brioude et al., 2018 (3)] and for the scoring system developed using the current cohort (0.88, 95% CI: 0.81–0.94, Figure 3). Table 2 shows the diagnostic efficacy of the scoring systems using clinical symptoms for epigenetically confirmed BWS.

Full table
Based on the differences in clinical symptoms between BWS epigenetic subtypes (Figure 2) and patient data from our cohort, we estimated that the AUC of ROC in the differentiation of one epigenetic subtype from the others was 0.78, 0.79, and 0.63 for IC2-LOM, uUPD, and IC1-GOM, respectively (Table 3).
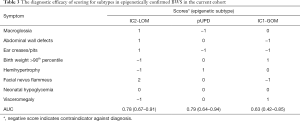
Full table
Discussion
The current study involved a cohort of patients who met our symptom-based diagnostic criteria for BWS that comprised all diagnostic criteria described to date. We found that the current clinical indicators for BWS had a positive prediction value of almost 70% for a positive epigenetic test result. The recently developed scoring systems for diagnosis had similar efficacy, which is convenient for the clinical diagnosis of patients with BWS. Clear differences were identified in clinical features between BWS subtypes, and these could serve as good indicators in the differentiation of BWS epigenetic subtypes. However, differences between European and Asian populations were not evident for most clinical features.
The majority of BWS studies to date have been conducted on European populations (12,14,16,18-24), and studies on Asian populations are few and have smaller sample sizes (17,25,27,28). Differences between European and Asian populations have been indicated in previous studies (1,15), as have differences within Asian populations. For instance, in Taiwan, the most common features of BWS were found to be macroglossia (74%), abdominal wall defects (63%), and ear creases or pits (58.5%) (27); in Hong Kong, visceromegaly (78%), macroglossia (70%), and ear creases/pits (67%) were the most common features (17); and in Shanghai, the most common features were abdominal wall defect (84%), macroglossia (58%), and ear creases/pits (45%) (28). The rarity of the disease magnifies the inconsistency in the results from individual retrospective cohorts, which could be susceptible to bias due to confounding factors created by differences in patient selection, healthcare routines, and symptom awareness. Indeed, heterogeneity among studies has commonly been detected in the distribution of disease characteristics in both European studies with larger sample size and smaller Asian studies. The lower frequency of neonatal hypoglycemia among Asians may be related to under diagnosis, owing to the lack of neonatal hypoglycemia screening programs (31). To obtain the true rate of hypoglycemia among Asian populations with BWS, it would be necessary to observe pediatric hypoglycemia screening and accumulate more data in future studies. Nevertheless, the current analysis suggested that the difference in BWS between populations might be too insignificant to be ascertained by studies on similar scale with confounding factors that cannot be easily controlled.
The aggregation of clinical features in BWS epigenetic subtypes could reflect the underlying mechanisms of the disease. For instance, IGF2 overexpression could explain the higher rate of body overgrowth and visceromegaly in BWS with IC1-GOM (32). Despite being the epigenetic combination of both IC1-GOM and IC2-LOM, the pUPD subtype has not been associated with the highest presentation of any clinical symptoms apart from hemihypertrophy, which is probably attributable to the commonly identified asymmetrical post mitotic mosaicism in patients with pUPD or other factors that affect gene expression in 11p15.5 in a tissue-specific manner (6,33,34). Nevertheless, the mechanisms of the abdominal wall defects and abnormal ear or facial appearance seen in patients with IC2-LOM are currently unclear.
Patients with BWS have increased risk of childhood cancer and, therefore, need regular surveillance (3,11). The difference in tumor occurrence among epigenetic subtypes (e.g., Wilms’ tumor in IC1-GOM and hepatoblastoma in pUPD) indicates a need for stratified follow-up programs for patients with BWS subtypes (11). Although the current evidence confirms the difference in clinical symptoms between epigenetic subtypes, our study also suggested that the diagnosis of BWS subtypes needs to be by genetic analysis to supplement categorization according to clinical symptoms (the AUCs of which were no more than 0.8).
Our ROC curve analysis showed that the BWS diagnostic scoring systems had similar efficacy. Although these diagnostic tools can show high specificity in patients who present with a more number of BWS-associated symptoms, they cannot effectively predict epigenetic-positive BWS in patients with fewer symptoms. The rarity of the disease limits the possibility for sensitive diagnostic criteria solely based on clinical symptoms. Improving the diagnosis of BWS might depend on increasing the accessibility to genetic testing, which would allow more patients to be confirmed using a reduced diagnostic cutoff (18).
One of the limitations of our study is its small sample size, which could partly explain our study’s deviation from the overall pattern in relation to clinical characteristics. Moreover, we did not conduct further genetic testing on patients who tested negative in the epigenetic test. The addition of tests for CDKN1C mutations (by sequencing), chromosomal rearrangement (by karyotyping), and low-level mosaicism (using MS-PCR of MS-qPCR) could result in more than 70% positive molecular results based on our inclusion criteria. The development of an (epi)genetic triage for symptomatic BWS patients based on the accessibility to molecular tests, as well as a strategy for identifying new pathogenic molecular mechanisms, would improve the diagnosis of the disease.
Conclusions
In conclusion, our analysis of a Chinese cohort of BWS patients confirmed the diagnostic efficacy of recently developed symptom-based BWS scoring systems, which was effective in the early identification of patients with BWS. There were no clear differences in most BWS symptoms between Asian and European populations. However, the differences in phenotypes between BWS epigenetic subtypes are significant.
Acknowledgments
We are grateful to the families who participated in the study.
Funding: The study was supported by the Children’s Hospital of Fudan University (no. EK112520180204).
Footnote
Reporting Checklist: The authors have completed the STROBE reporting checklist. Available at http://dx.doi.org/10.21037/tp-20-243
Data Sharing Statement: Available at http://dx.doi.org/10.21037/tp-20-243
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/tp-20-243). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). Informed consent was acquired by parents of all the patients before enrollment. This study was approved by the Medical Ethics Committee of the Children’s Hospital of Fudan University.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Barisic I, Boban L, Akhmedzhanova D, et al. Beckwith Wiedemann syndrome: A population-based study on prevalence, prenatal diagnosis, associated anomalies and survival in Europe. Eur J Med Genet 2018;61:499-507. [Crossref] [PubMed]
- Mussa A, Russo S, De Crescenzo A, et al. Prevalence of Beckwith-Wiedemann syndrome in North West of Italy. Am J Med Genet A 2013;161A:2481-6. [PubMed]
- Brioude F, Kalish JM, Mussa A, et al. Expert consensus document: Clinical and molecular diagnosis, screening and management of Beckwith-Wiedemann syndrome: an international consensus statement. Nat Rev Endocrinol 2018;14:229-49. [Crossref] [PubMed]
- Choufani S, Shuman C, Weksberg R. Beckwith-Wiedemann syndrome Am J Med Genet C Semin Med Genet 2010;154C:343-54. [Crossref] [PubMed]
- Cerrato F, Vernucci M, Pedone PV, et al. The 5' end of the KCNQ1OT1 gene is hypomethylated in the Beckwith-Wiedemann syndrome. Hum Genet 2002;111:105-7. [Crossref] [PubMed]
- Romanelli V, Meneses HN, Fernández L, et al. Beckwith-Wiedemann syndrome and uniparental disomy 11p: fine mapping of the recombination breakpoints and evaluation of several techniques. Eur J Hum Genet 2011;19:416-21. [Crossref] [PubMed]
- Wang KH, Kupa J, Duffy KA, et al. Diagnosis and Management of Beckwith-Wiedemann Syndrome. Front Pediatr 2020;7:562. [Crossref] [PubMed]
- Munns CF, Batch JA. Hyperinsulinism and Beckwith-Wiedemann syndrome Arch Dis Child Fetal Neonatal Ed 2001;84:F67-9. [Crossref] [PubMed]
- DeBaun MR, King AA, White N. Hypoglycemia in Beckwith-Wiedemann syndrome. Semin Perinatol 2000;24:164-71. [Crossref] [PubMed]
- Angelousi A, Kyriakopoulos G, Nasiri-Ansari N, et al. The role of epithelial growth factors and insulin growth factors in the adrenal neoplasms. Ann Transl Med 2018;6:253. [Crossref] [PubMed]
- Maas SM, Vansenne F, Kadouch DJ, et al. Phenotype, cancer risk, and surveillance in Beckwith-Wiedemann syndrome depending on molecular genetic subgroups. Am J Med Genet A 2016;170:2248-60. [Crossref] [PubMed]
- Brioude F, Lacoste A, Netchine I, et al. Beckwith-Wiedemann syndrome: growth pattern and tumor risk according to molecular mechanism, and guidelines for tumor surveillance. Horm Res Paediatr 2013;80:457-65. [Crossref] [PubMed]
- Trobaugh-Lotrario AD, Venkatramani R, Feusner JH. Hepatoblastoma in children with Beckwith-Wiedemann syndrome: does it warrant different treatment? J Pediatr Hematol Oncol 2014;36:369-73. [Crossref] [PubMed]
- Zarate YA, Mena R, Martin LJ, et al. Experience with hemihyperplasia and Beckwith-Wiedemann syndrome surveillance protocol. Am J Med Genet A 2009;149A:1691-7. [Crossref] [PubMed]
- Duffy KA, Sajorda BJ, Yu AC, et al. Beckwith-Wiedemann syndrome in diverse populations. Am J Med Genet A 2019;179:525-33. [PubMed]
- Mussa A, Russo S, De Crescenzo A, et al. (Epi)genotype-phenotype correlations in Beckwith-Wiedemann syndrome. Eur J Hum Genet 2016;24:183-90. [Crossref] [PubMed]
- Luk HM. Clinical and molecular characterization of Beckwith-Wiedemann syndrome in a Chinese population. J Pediatr Endocrinol Metab 2017;30:89-95. [Crossref] [PubMed]
- Ibrahim A, Kirby G, Hardy C, et al. Methylation analysis and diagnostics of Beckwith-Wiedemann syndrome in 1,000 subjects. Clin Epigenetics 2014;6:11. [Crossref] [PubMed]
- Weksberg R, Shuman C, Beckwith JB. Beckwith-Wiedemann syndrome. Eur J Hum Genet 2010;18:8-14. [Crossref] [PubMed]
- Cooper WN, Luharia A, Evans GA, et al. Molecular subtypes and phenotypic expression of Beckwith-Wiedemann syndrome. Eur J Hum Genet 2005;13:1025-32. [Crossref] [PubMed]
- Elliott M, Bayly R, Cole T, et al. Clinical features and natural history of Beckwith-Wiedemann syndrome: presentation of 74 new cases. Clin Genet 1994;46:168-74. [Crossref] [PubMed]
- DeBaun MR, Tucker MA. Risk of cancer during the first four years of life in children from The Beckwith-Wiedemann Syndrome Registry. J Pediatr 1998;132:398-400. [Crossref] [PubMed]
- Gaston V, Le Bouc Y, Soupre V, et al. Analysis of the methylation status of the KCNQ1OT and H19 genes in leukocyte DNA for the diagnosis and prognosis of Beckwith-Wiedemann syndrome. Eur J Hum Genet 2001;9:409-18. [Crossref] [PubMed]
- Weksberg R, Nishikawa J, Caluseriu O, et al. Tumor development in the Beckwith-Wiedemann syndrome is associated with a variety of constitutional molecular 11p15 alterations including imprinting defects of KCNQ1OT1. Hum Mol Genet 2001;10:2989-3000. [Crossref] [PubMed]
- Lee BH, Kim GH, Oh TJ, et al. Quantitative analysis of methylation status at 11p15 and 7q21 for the genetic diagnosis of Beckwith-Wiedemann syndrome and Silver-Russell syndrome. J Hum Genet 2013;58:604-10. [Crossref] [PubMed]
- Viberti G. Recent advance in the treatment of diabetic nephropathy: low-protein diet. J Diabet Complications 1991;5:87. [Crossref] [PubMed]
- Lin HY, Chuang CK, Tu RY, et al. Epigenotype, genotype, and phenotype analysis of patients in Taiwan with Beckwith-Wiedemann syndrome. Mol Genet Metab 2016;119:8-13. [Crossref] [PubMed]
- Wang R, Xiao Y, Li D, et al. Clinical and molecular features of children with Beckwith-Wiedemann syndrome in China: a single-center retrospective cohort study. Ital J Pediatr 2020;46:55. [Crossref] [PubMed]
- Duffy KA, Cielo CM, Cohen JL, et al. Characterization of the Beckwith-Wiedemann spectrum: Diagnosis and management. Am J Med Genet C Semin Med Genet 2019;181:693-708. [Crossref] [PubMed]
- Viechtbauer W. Conducting Meta-Analyses in R with the metafor Package. J Stat Softw 2010;36:1-48. [Crossref]
- Thompson-Branch A, Havranek T. Neonatal Hypoglycemia. Pediatr Rev 2017;38:147-57. [Crossref] [PubMed]
- Leighton PA, Ingram RS, Eggenschwiler J, et al. Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 1995;375:34-9. [Crossref] [PubMed]
- Ohtsuka Y, Higashimoto K, Oka T, et al. Identification of consensus motifs associated with mitotic recombination and clinical characteristics in patients with paternal uniparental isodisomy of chromosome 11. Hum Mol Genet 2016;25:1406-19. [Crossref] [PubMed]
- Urzua A, Burattini S, Pinochet C, et al. Somatic Mosaicism for Paternal Uniparental Disomy of 11p15.5 Region in Adrenal and Liver Tissues in a Newborn with Atypical Beckwith-Wiedemann Syndrome. J Pediatr Genet 2019;8:226-30. [Crossref] [PubMed]
(English Language Editor: J. Reynolds)


