
Ras pathway mutation feature in the same individuals at diagnosis and relapse of childhood acute lymphoblastic leukemia
Introduction
Acute lymphoblastic leukemia (ALL) is the most common malignancy in children (1,2). The prognosis of children with ALL has been greatly improved in recent years due to the precise classification based on MICM (morphology, immunology, cytogenetics, and molecular biology), the application of combined chemotherapy and the use of molecular targeted drugs. The 5-year overall survival rate (OS) is approximately 90%; however, 20% of patients suffer ALL relapse, resulting in treatment failure and even death (2-5). The relapse of ALL remains a leading cause of death in children.
Molecular alterations are common in ALL patients, and some molecular features, such as abnormal chromosome structure and number, fusion genes, and gene mutations, have become the basis for ALL classification and prognosis prediction. The development of next-generation sequencing (NGS) technology has revealed more abnormal molecules and improved the understanding of ALL. Some abnormal molecules associated with chemotherapy resistance and relapse were observed in patients with ALL relapse, such as the resistance of CREBBP mutations to glucocorticoids (6-8), NT5C2 mutations to 6-mercaptopurine (9-11), and PRPS1 mutations to 6-mercaptopurine (12). The deletion of the NS3C1 gene was observed in TEL-AML1 positive patients with ALL relapse (13). Ras pathway mutations in genes such as KRAS, NRAS, FLT3, and PTPN11 were observed in more than 50% patients with ALL relapse (5,14), resulting in chemotherapy resistance to prednisone.
Like many malignancies, ALL has clonal diversity. Chemotherapeutic drugs can induce subclonal mutations, leading to drug resistance. Analysis of the same patients at the initial diagnosis and relapse revealed that relapse-acquired mutations mainly participated in epigenetic regulation, tumor suppression, Ras signal transduction and drug metabolism (6-12,15). Clonal lineage changes from initial diagnosis to relapse were constructed, indicating that the heterogeneity of the clones was very common in ALL patients. Primary clones at the time of ALL relapse usually evolved from secondary clones that existed at the time of initial diagnosis (15,16). Therefore, the analysis of clonal evolution in the same pediatric patients from the initial diagnosis to relapse and the exploration of the main reason for ALL relapse can improve the understanding of the underlying molecular characteristics, and guide the study of the pathogenesis of ALL.
In this study, we aimed to use NGS technology to analyze gene alterations during the initial diagnosis and relapse of childhood ALL, and to explore the main reasons leading to ALL relapse.
Methods
Ethical compliance
Informed consent was obtained from the patients’ legal guardians in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of children’s hospital of Fudan University (No. [2015]005), Shanghai, China.
Patients and samples
The study included 12 Chinese pediatric patients with ALL, who had been primary diagnosed and treated in the children’s hospital of Fudan University, and suffered ALL relapse between January 2015 and December 2017. The diagnosis of ALL was based on the World Health Organization’s classification and patients were treated with modified St.Jude XV protocol (17). Bone marrow samples at the time of primary diagnosis and relapse were collected, and matched bone marrow samples at remission were used as germline controls.
Patients’ basic information (including age, sex, etc.), clinical manifestations, laboratory examinations and treatment events were retrospectively collected and analyzed. Laboratory examinations included morphological, immunophenotyped and cytogenetic analyses, peripheral blood examination, cerebrospinal fluid examination, and imaging tests for testicular invasion.
Assessment of response to treatment
Complete remission (CR) was defined as the presence of less than 5% blasts in the bone marrow on day 46 of induction chemotherapy, and no evidence of extramedullary disease. Minimal residual disease (MRD) analysis was recommended as combined risk stratification using flow cytometry. Relapse was defined as the reappearance of leukemic cells in bone marrow with >25% blasts. Central nervous system relapse was defined as more than five blasts in the cerebrospinal fluid. Testicular relapse was diagnosed clinically and confirmed with ultrasonography. Very early relapse was defined that overall time was less than 18 months after initial diagnosis; early relapse was defined that overall time was more than 18 months after initial diagnosis but less than 6 months after chemotherapy finished; late relapse was more than 6 months after the end of chemotherapy (18).
Targeted capture sequencing and mutation analysis
Genomic DNA was extracted from cell pellets using the DNAeasy Blood and Tissue Kit (Qiagen, USA) according to the experimental manipulate. DNA was quantified using a Qubit Fluorometer (Life Technologies, USA), and DNA integrity was assessed by agarose gel electrophoresis. Methods for gene sequencing and data analysis were based on our published studies (19).
Statistical analysis
SPSS 24.0 (SPSS, Chicago) was used for statistical analysis. Continuous variables were compared using t-test or non-parametric tests; categorical variables were compared by Pearson’s Chi square test or Fisher’s exact test. Two-sided P<0.05 was considered statistically significant.
Results
Clinical features of patients
There were 12 patients with ALL relapse, including 6 males and 6 females; four cases of T-cell acute lymphoblastic leukemia (T-ALL) and 8 cases of pre-B-cell acute lymphoblastic leukemia (B-ALL). The median diagnosis age was 5.8 years (0.8–11.8 years), and the median number of white blood counts (WBCs) was 52.1×109/L (0.5–445.1×109/L). Four cases were fusion gene-positive, including 1 TEL-AML1 case, 1 SIL-TAL1 case, and 2 MLLr (MLL rearrangement) cases. In patients with abnormal chromosome numbers, one patient had hyperdiploidy (number of chromosomes ≥50), and one patient had hypodiploidy (number of chromosomes <40). Among the 12 patients, 2 were at low risk (LR), 9 were at intermediate risk (IR), and 1 was at high risk (HR).
All patients with ALL relapse achieved CR in terms of bone marrow at the early stage of treatment (at the 46th day of the treatment, the number of primitive naive cells in bone marrow was <5%), and 10 cases (83.3%) had MRD values less than 0.01%. Among the 12 patients, 8 patients experienced very early relapse, and the main site of relapse was bone marrow. Table 1 shows the clinical features of the 12 patients with ALL relapse.
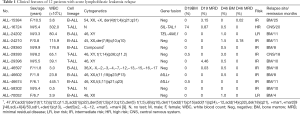
Full table
Analysis of clonal evolution
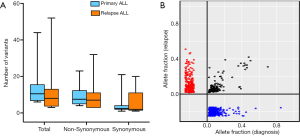
Paired analysis of the same 12 pediatric patients at the initial diagnosis and relapse of ALL showed that the average number of mutations was 15 (range, 1–44) at the initial diagnosis, and 13 (range, 0–52) at relapse. Though the initial diagnosis had more somatic mutations, no significant differences were observed (P=0.732), while changes in gene clones occurred after relapse (Figure 1). Seven ALL patients (58.3%) presented with clones of a small number of genes at the initial diagnosis (Figure 2). Overall, the clones of the harmful mutations in the same patient changed significantly (with the exception of ALL-46602 and ALL-58302), and the overall change was that during relapse, some clones were lost, some clones were acquired, some clones increased, and some clones decreased.
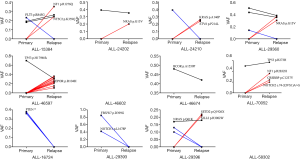
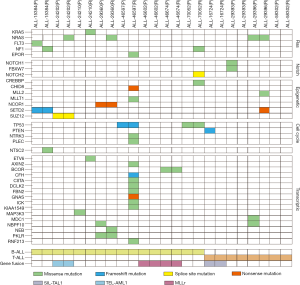
Gene alteration
At initial diagnosis, the number of mutations in the 4 T-ALL patients was similar to that of the 8 B-ALL patients (2.25 vs. 2.13, P=0.914). However, at the time of relapse, the number of mutations in T-ALL patients was less than that of B-ALL patients [on average 0.75 mutations (range, 0–3) vs. 4.25 mutations (range, 0–16)], while the difference was not significant (P=0.213). In addition, one T-ALL patient (ALL-58302) and one B-ALL (ALL-46002) patient did not have any target mutations at both the time of initial diagnosis and relapse. Moreover, at the initial diagnosis, 4 fusion gene-positive patients had an average of 1.5 harmful mutations (range, 0–4), and fusion gene-negative patients had an average of 2.5 mutations (range, 0–6), while there was no significant difference between them (P=0.376). One MLLr patient (ALL-46602) did not carry any mutations, and 1 SIL-TAL1 positive patient (ALL-16724) had no mutations at the time of relapse.
Gene alterations in the Ras signaling pathway
Among the 8 B-ALL patients, 6 (75%) patients obtained gene alterations (NRAS, KRAS, NF1, and EPOR) in the Ras pathway during relapse, including 2 cases (ALL-24202, ALL-29360) with the NRAS p.G13V mutation, 1 case (ALL-24210) with the KRAS p.A146P mutation, 2 cases (ALL-15384, ALL-70052) with NF1 mutations (p.R1276Q and p.R2832H), and 1 case (ALL-46597) with the EPOR p.R104H mutation; Among the 4 T-ALL patients, 1 patient (ALL-29396) had the NRAS p.Q61K mutation at both the time of initial diagnosis and relapse. Gene alterations in the Ras pathway are more common in patients with B-ALL than in patients with T-ALL during relapse (Figures 2,3).
Gene alterations in the Notch signaling pathway
Among the 4 T-ALL patients, 1 patient (ALL-29393) had both the NOTCH1 p.D399G and FBXW7 p.L1678P mutations at initial diagnosis but had neither the NOTCH1 p.D399G nor FBXW7 p.L1678P mutation or any other gene alteration at relapse. One patient with B-ALL (ALL-70052) acquired the NOTCH2 c.74-2(IVS1)A>G mutation at the time of relapse (Figures 2,3).
Epigenetic regulation-associated gene alterations
One ALL patient (ALL-29396) had the MLL2 p.R3082W mutation, a newly acquired mutation site, which was predicted to be harmful by the protein structure. Two ALL patients had SETD2 mutations: one (ALL-29396) had a truncating mutation (p.Q1926X) during relapse, and the other (ALL-15384) had frameshift mutations at 2 sites (p.S165Lfs*12,p.A158Dfs*13) in SETD2, both at the time of the initial diagnosis and relapse; the occurrence of either frameshift mutations or truncating mutations leads to functional loss of the SETD2 gene. One B-ALL patient (ALL-70052) acquired the CREBBP p.C1237Y mutation and other mutations, including the SUZ12 c.1875-1(IVS15)G>A, CHD8 p.R2044X, MLLT1 p.A17T and NCOR1 p.R350X mutation, during relapse (Figures 2,3).
Cell cycle-associated gene alterations
Two B-ALL patients (ALL-46597 and ALL-70052) had TP53 mutations (p.H179Mfs and p.R273H) with VAFs greater than 30%, indicating changes in the primary clones. One T-ALL (ALL-16724) had mutations at multiple sites in PTEN at the time of initial diagnosis but had no such mutations at the time of relapse. Another B-ALL (ALL-46597) acquired the NTRK3 p.R507H and PLEC p.G4456R mutations during relapse (Figures 2,3).
Other gene alterations
NT5C2 is a relapse-associated gene, and we only observed the NT5C2 p.K359Q mutation in 1 patient with B-ALL (ALL-15384) (Figures 2,3).
Discussion
Like many malignancies, ALL has clonal diversity. Clones of genes undergo major changes from the time of initial diagnosis to the time of relapse. Approximately 58.3% of the relapsed patients had clones from the initial diagnosis, and the same gene could have multiple subclonal changes at the same time, including the increase and decrease in clones at the time of relapse, and subclones can change to primary clones during relapse. These results support the nonlinear evolution in malignancies, especially in leukemia. Clones are mainly evolved from branches in relapsed ALL. These branches are derivatives of the ancestral subclone that are associated with but different from other major leukemic clones at the initial diagnosis (14). These mutations play a driving role and may be the second event in the occurrence and development of leukemia. Further, chemotherapy itself may induce drug mutations, chemotherapy-induced resistance mutations may occur through two-step-selection of a persister clone, which survives initial treatment and later gives rise to the true resistant clones (20,21).Thus, in most cases the predominant clones present at diagnosis were eradicated by therapy, further studies should use sensitive mutation testing to examine the relationship between the nature of the therapeutic regimen and the mutational specture.
Recently, several genetic defects have been linked to chemotherapy resistance in relapse ALL, including CREBBP, NT5C2, TP53, PRPS1, PRPS2, WHSC1, NR3C1, MSH2, MSH6, PMS2, FPGS, causing resistance to various drugs (6,9,10,21). NT5C2 and PRPS1 were not detectable at diagnosis even with high-depth sequencing, suggesting these mutations were acquired after diagnosis and possibly during treatment (21). In our cases, only one case occurred NT5C2 or CREBBP mutation at relapse. The NT5C2 gene encodes a cytoplasmic nucleotidase that is involved in the clearance of cytotoxic metabolites of 6-mercaptopurine and 6-thioguanine. Resistance to 6-mercaptopurine was observed in 20% of T-ALL patients and 5% of B-ALL patients (6,9,10). The NT5C2 p.K359Q site is a known drug-resistance site. In a CCRF-CEM cell experiment, the 6-MP IC50 was 6.47 µM while the wild type IC50 was 3.46 µM, and the 6-MP activity was significantly reduced, affecting drug metabolism (9,10). CREBBP, which encodes the cAMP-response element-binding protein, acts as a transcription cofactor that participates in multiple signaling pathways. CREBBP mutations are common in patients with hyperdiploidy, and patients with KRAS mutations are prone to early relapse, and acquired mutations increase the risk of ALL relapse. The CREBBP p.C1237Y mutation affects Histone acetyltransferase (HAT) domain, reduces acetylation of histone and nonhistone proteins, and reduces cell proliferation, leading to glucocorticoid resistance (7,8). CREBBP mutations can enhance gene mutations in the Ras pathway in ALL patients, but MEK inhibitors are effective for ALL patients with Ras pathway mutations and with or without CREBBP mutations (22). Thus, specific therapeutic agents should be considered carefully, including nucleoside metabolism analogues (NT5C2) and glucocorticoids (CREBBP), alternative treatment approaches to avoid mutation selection or mitigate resistance may be needed.
Our results also showed that there were 7 patients (58%) had mutations in the Ras pathway (KRAS, NRAS, NF1, EPOR, etc.) among the 12 patients and 75% B-ALL patients obtained gene alterations during relapse. Previous studies showed that Ras mutations could be observed in 30–50% of patients with relapsed ALL (4,5,8,14,23,24). These observations confirm that Ras pathway mutations are frequent at relapse in pediatric B-ALL. Nearly all mutations in the Ras signaling pathway at diagnosis were acquired after emergency of founder clone. The clonal evolution of Ras mutations is highly heterogeneous. Some patients retain the original mutant clones during relapse, some lose original clones, and some patients acquire or lose new mutant clones during treatment. Remarkably, 5 out of 8 B-ALL cases gained clonal mutations of Ras pathway at relapse (VAF more than 25%), which is different to Jerchel’s research that clonal mutations at initial diagnosis were retained at relapse and subclonal mutations often expanded at relapse (24). Ras mutations are often subclonal changes, induced by chemotherapeutic drugs in animal experiments may lead to drug resistance to dexamethasone but are sensitive to vincristine, leading to positive or negative Ras mutations during relapse (6,8). However, the prognosis of subclonal mutations in Ras pathway was similar to wild-type cases, so subclonal mutations should not be assigned to a higher risk group or treatment with MEK inhibitor (24). Furthermore, NRAS and KRAS mutations are associated with a poorer overall survival (23,25), targeted MEK inhibition may offer clinical benefit for a substantial proportion of children with relapsed ALL and MEK inhibition can synergize with prednisolone (24).
In addition, MLLr is well-recognized for its poor prognosis and easy to occur relapse early; TP53 mutations mostly occur in low hypodiploid (chromosome <44) and are observed in both newly diagnosed and relapsed ALL, approximately 50% of which were germline mutation in nature (6,26). Children with TP53 variants were at a higher risk of ALL or second cancers (27). In our previous study, MLLr and TP53 mutations were powerful predictor for an adverse outcome in B-ALL and pediatric ALL (19).
There are some limitations to the present study. We noticed a few patients with high mutational burden at diagnosis, but did not have any mutations at relapse, for example ALL-16724 and ALL-29393, the possible reason was low proportion of blast cells in the bone marrow and technology limitation-based tumor purity may cause underestimation of clonal diversity due to failure in identifying low levels of mutations clusters. Sensitive mutation detection strategies at multiple time points during diagnosis-therapy-relapse are needed to detect low levels of mutant clone and to examine the relationship between the therapy regimen and the mutational specture in the further study. Also, the limited number of cases in our study lead to the analysis of the clones was limited, and large scale sample and the optimal interval for monitoring ALL-specific mutations before relapse by high-depth sequencing of serial bone marrow sample are needed to reveal the characteristics of molecular alterations in childhood ALL, guide clinical decision-making, and achieve precise classification and treatment.
Conclusions
In conclusion, NGS technology, based on its characteristics of high throughput and high sensitivity and specificity, provides a good platform for the clinical study of ALL and improves the understanding of molecular alterations in ALL patients. Paired ALL samples of initial diagnosis and relapse were subjected to NGS, not only it could reveal the key mutations that lead to relapse, but also we can study the evolutionary lineage and understand the key issues in the evolutionary process of tumor clones.
Acknowledgements
We thank all patients and families who participated in this study.
Funding: The research was funded by the Research Programs of the Shanghai Science and Technology Commission Foundation (No.14411950603), Shanghai Municipal Commission of Health and Family Planning (No.201740011). Project Ai You Foundation Supporting Children with Cancer Program.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Informed consent was obtained in accordance with the Declaration of Helsinki and approved by the Institutional Review Board of Children’s Hospital of Fudan University (No. [2015]005), Shanghai, China.
References
- Inaba H, Greaves M, Mullighan CG. Acute lymphoblastic leukemia. Lancet 2013;381:1943-55. [Crossref] [PubMed]
- Yu H, Du Y, Xu J, et al. Prognostic relevance of genetic variations in T-cell acute lymphoblastic leukemia/lymphoblastic lymphoma. Transl Cancer Res 2019;8:2485-95. [Crossref]
- Chiaretti S, Gianfelici V, Ceglie G, et al. Genomic Characterization of Acute Leukemia. Med Princ Pract 2014;23:487-506. [Crossref] [PubMed]
- Ding ZC. The promise and challenges of chimeric antigen receptor T cells in relapsed B-cell acute lymphoblastic leukemia. Ann Transl Med 2018;6:235. [Crossref] [PubMed]
- Tasian SK, Hunger SP. Genomic characterization of pediatric acute lymphoblastic leukemia: an opportunity for precision medicine therapeutics. Br J Haematol 2017;176:867-82. [Crossref] [PubMed]
- Ma X, Edmonson M, Yergeau D, et al. Rise and fall of subclones from diagnosis to relapse in pediatric B-acute lymphoblastic leukemia. Nat Commun 2015;6:6604. [Crossref] [PubMed]
- Mullighan CG, Zhang J, Kasper LH, et al. CREBBP mutations in relapsed acute lymphoblastic leukemia. Nature 2011;471:235-9. [Crossref] [PubMed]
- Malinowska-Ozdowy K, Frech C, Schönegger A, et al. KRAS and CREBBP mutations: a relapse-linked malicious liaison in childhood high hyperdiploid acute lymphoblastic leukemia. Leukemia 2015;29:1656-67. [Crossref] [PubMed]
- Tzoneva G, Perezgarcia A, Carpenter Z, et al. Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL. Nat Med 2013;19:368-71. [Crossref] [PubMed]
- Meyer JA, Wang J, Hogan LE, et al. Relapse-specific mutations in NT5C2 in childhood acute lymphoblastic leukemia. Nat Genet 2013;45:290-4. [Crossref] [PubMed]
- Mar BG, Bullinger LB, McLean KM, et al. Mutations in epigenetic regulators including SETD2 are gained during relapse in paediatric acute lymphoblastic leukaemia. Nat commun 2014;5:3469. [Crossref] [PubMed]
- Li B, Li H, Bai Y, et al. Negative feedback-defective PRPS1 mutants drive thiopurine resistance in relapsed childhood ALL. Nat Med 2015;21:563-71. [Crossref] [PubMed]
- Kuster L, Grausenburger R, Fuka G, et al. ETV6/RUNX1-positive relapses evolve from an ancestral clone and frequently acquire deletions of genes implicated in glucocorticoid signaling. Blood 2011;117:2658-67. [Crossref] [PubMed]
- Oshima K, Khiabanian H, da Silva-Almeida AC, et al. Mutational landscape, clonal evolution patterns, and role of RAS mutations in relapsed acute lymphoblastic leukemia. Proc Natl Acad Sci U S A 2016;113:11306-11. [Crossref] [PubMed]
- Mullighan CG, Phillips LA, Su X, et al. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science 2008;322:1377-80. [Crossref] [PubMed]
- Lindqvist CM, Lundmark A, Nordlund J, et al. Deep targeted sequencing in pediatric acute lymphoblastic leukemia unveils distinct mutational patterns between genetic subtypes and novel relapse-associated genes. Oncotarget 2016;7:64071-88. [Crossref] [PubMed]
- Pui CH, Campana D, Pei D, et al. Treating childhood acute lymphoblastic leukemia without cranial irradiation. N Engl J Med 2009;360:2730-41. [Crossref] [PubMed]
- Einsiedel HG, von Stackelberg A, Hartmann R, et al. Long-term outcome in children with relapsed ALL by risk-stratified salvage therapy: results of trial acute lymphoblastic leukemia-relapse study of the Berlin-Frankfurt-Munster Group 87. J Clin Oncol 2005;23:7942-50. [Crossref] [PubMed]
- Zhang HH, Wang HS, Qian XW, et al. Genetic variants and clinical significance of pediatric acute lymphoblastic leukemia. Ann Transl Med 2019. [Crossref] [PubMed]
- Sharma SV, Lee DY, Li B, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010;141:69-80. [Crossref] [PubMed]
- Li B, Brady SW, Ma X, et al. Therapy-induced mutations drive the genomic landscape of relapsed acute lymphoblastic leukemia. Blood 2020;135:41-55. [PubMed]
- Dixon ZA, Nicholson L, Zeppetzauer M, et al. CREBBP knockdown enhances RAS/RAF/MEK/ERK signaling in Ras pathway mutated acute lymphoblastic leukemia but does not modulate chemotherapeutic response. Haematologica 2017;102:736-45. [Crossref] [PubMed]
- Irving J, Matheson E, Minto L, et al. Ras pathway mutations are prevalent in relapsed childhood acute lymphoblastic leukemia and confer sensitivity to MEK inhibition. Blood 2014;124:3420-30. [Crossref] [PubMed]
- Jerchel IS, Hoogkamer AQ, Ariës IM, et al. Ras pathway mutations as a predictive biomarker for treatment adaptation in pediatric B-cell precursor acute lymphoblastic leukemia. Leukemia 2018;32:931-40. [Crossref] [PubMed]
- Irving JA, Enshaei A, Parker CA, et al. Integration of genetic and clinical risk factors improves prognostication in relapsed childhood B-cell precursor acute lymphoblastic leukemia. Blood 2016;128:911-22. [Crossref] [PubMed]
- Holmfeldt L, Wei L, Diazflores E, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet 2013;45:242-52. [Crossref] [PubMed]
- Qian M, Cao X, Devida M, et al. TP53 Germline Variations Influence the Predisposition and Prognosis of B-Cell Acute Lymphoblastic Leukemia in Children. J Clin Oncol 2018;36:591-9. [Crossref] [PubMed]


