Uniparental disomy in Robertsonian translocations: strategies for uniparental disomy testing
Concept of uniparental disomy (UPD)
UPD as a concept was first introduced by Engel in 1980 and was defined as the inheritance or presence in a diploid offspring of both homologs of a pair of chromosomes from one parent only with no contribution from the second parent (1). Depending on the origin of the disomic chromosome, UPD is denoted as maternal or paternal.
UPD can be further classified as heterodisomy (hUPD) or isodisomy (iUPD). In full hUPD the two inherited chromosomes represent a chromosome pair from a single parent. In iUPD there are two identical copies of one of the two parental chromosomes. Segmental UPD has also been demonstrated by molecular analyses to involve a region of both chromosomes of a pair with the rest of the chromosome pair biparentally inherited. UPD in Man was first demonstrated for maternal iUPD 7 in a patient with cystic fibrosis and short stature (2). Both homologs of chromosome 7 were identical and were derived from the mother.
UPD has been identified in investigations of prenatal and postnatal chromosomal mosaicism; recessive genetic disease; structurally abnormal rearrangements and phenotypes associated with imprinting (3). UPD is generally demonstrable at the molecular level and in 65% of known UPD cases, presents as a cytogenetically normal 46,XX or 46,XY karyotype (4). Currently, over 2,500 UPD cases have been documented in a regularly updated online database which centres on UPD in clinically normal and abnormal individuals who have normal or abnormal karyotypes (5). Abnormal karyotypes can reflect rearrangements that are either balanced or unbalanced. Balanced rearrangements associated with UPD include Robertsonian translocations (ROBs), balanced reciprocal translocations, isochromosomes and inversions and these represent at least 8% of all UPD cases reported (4). Unbalanced rearrangements like small supernumerary marker chromosomes, unbalanced reciprocal translocations, and partial deletions and duplications were estimated to be involved in over 16% of UPD cases (4).
ROBs and the incidence of UPD
ROBs are among the most common balanced structural rearrangements seen with an incidence in the human population of 1 in 1,000 (6). They result from whole arm exchanges of the five human acrocentric chromosomes (chromosomes 13, 14, 15, 21 and 22). This type of rearrangement was named after the American biologist William Robertson who first described the fusion of two acrocentrics in grasshoppers in 1916. These ROBs are considered balanced and carriers with loss of the short arms have 45 chromosomes. The ROBs can be dicentric involving two centromeres but may appear monocentric with a “suppressed” centromere (7). Located in the stalks of the short arms of all five acrocentrics are multiple copies of the 18S and 28S ribosomal RNA genes with tandem satellite DNA repeats. No adverse phenotypic significance is attached to the loss of the short arms. The majority of ROBs (90%) involve non-homologous chromosomes with rob(13q;14q) the most common (8). True homologous ROBs involve the long arms of both homologs of the one acrocentric. Molecular studies using highly polymorphic markers can differentiate them from an acrocentric derived isochromosome from a single parental homolog. Some homologous ROBs and all isochromosomes will display uniparental inheritance (9).
The incidence of UPD of any chromosome is estimated to be 1:3,500 live births (10). Around 50% of the cases presented in the UPD online database (5) are associated with acrocentric chromosomes and over 10% of these acrocentric derived UPDs involve a Robertsonian tranlocation (4). An ascertainment bias not withstanding, most commonly documented in this database was UPD for chromosome 15, reflecting the many published reports of Prader Willi Syndromes (PWS) and Angelman syndromes (AS). It has been observed that around 30% of PWS are associated with UPD 15mat and 2-5% of AS with UPD 15pat (11,12).
It is of interest that one of the earliest established reports of UPD in PWS was associated with a maternally inherited rob(13q;15q) (13). Early UPD implications for chromosome 14 both maternal and paternal and described in 1991 were also derived from ROBs and involved rob(13q;14q) (14,15). A review of six postnatal studies of balanced ROBs with an abnormal phenotype, published between 1994 to 2000, revealed eighty-five non-homologous ROBs, four (4.7%) of these were UPD positive (16). Of the six homologous ROBs presenting with a normal phenotype, two uniparental cases were found to be derived from chromosomes 21 and 22.
Meiotic behaviour of a nonhomologous ROB
A nonhomologous (heterologous) ROB chromosome is comprised of the long arms of two different acrocentrics. Such a heterozygote with a ROB translocation chromosome and two normal acrocentric homologs would synapse as a trivalent at meiosis (Figure 1). Subsequent 2:1 segregation of a trivalent produces six types of gametes, two of which are normal and balanced resulting from an “alternate” segregation. Adjacent segregation however leads to two types of disomic and two types of nullisomic gametes. Very rarely would a 3:0 segregation in a ROB occur.

Semen analysis showed that 76-89% of spermatozoa were normal or balanced due to “alternate” segregation while nullisomics outnumber disomics amongst the unbalanced forms (17). Polar body analysis on rob(13q;14q) and rob(14q;21q) carriers showed unbalanced forms in the female to average around 33% and 42% respectively (18).
Mechanisms of UPD
The main mechanisms through which UPD may arise in ROBs include trisomy rescue, monosomy rescue and gamete complementation. Amongst reported UPD cases in the literature, trisomy rescue is the most common mechanism resulting in UPD. In trisomy rescue, a disomic gamete is produced by non-disjunction in one of the parents (most often maternal) and its fertilization by a normal haploid gamete would contribute to a trisomic conceptus. Loss of a homolog would result in UPD in 1/3 of the cases. It is recognised that most disomic gametes result from maternal meiosis I non-disjunction and therefore maternal hUPD would be the most common UPD resulting from such a rescue (19). Cases of UPD 15mat in PWS associated with placental mosaicism for trisomy 15 ascertained at CVS (chorion villus sampling) for advanced maternal age and subsequent correction of the trisomy have been well documented (20,21). More than 82% of the extra chromosomes in maternal UPD 15 have been associated with meiosis I (MI) non-disjunction errors and this would be consistent with the more commonly observed hUPD seen in maternal UPD (19). The study also found a lack of MI errors in males. Generally, paternal UDP 15 cases associated with AS reflect iUPD as a consequence of a postzygotic mitotic error though also possibly errors at meiosis II (MII) (19). Aneuploidy in human sperms is about five times less common than aneuploidy in oocytes (22).
ROB carriers are at increased risk of aneuploid offspring and are equally subject to trisomy rescue. The two more often noted mechanisms of trisomic and monosomic rescue generally involve two independent chromosome non-disjunction events. Using the example of an unbalanced adjacent segregation of a Robertsonian trivalent at maternal meiosis, disomic or nullisomic gametes when produced and fertilised by a normal haploid sperm would lead to a trisomic or monosomic zygote. A second event to restore the conception to a disomic state would require the loss of the extra chromosome in a trisomy as in trisomy rescue or the duplication of the single chromosome in a monosomy for monosomy rescue. Trisomy involving a Robertsonain translocation and its rescue through loss of a chromosome by resolving into a disomy would result in UPD in 50% of cases.
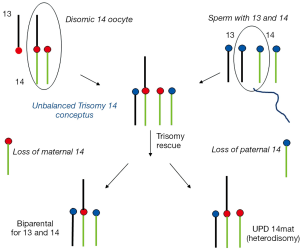
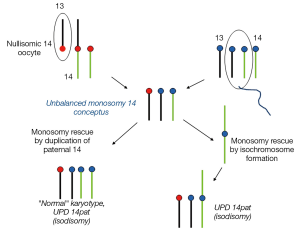
Examples of a rob(13;14) carrier mother giving rise to a UPD 14mat and UPD 14pat conceptus are outlined in Figures 2,3. The unbalanced trisomic 14 conceptus is “rescued” and resolved to a hUPD (UPD 14mat) by loss of the paternal 14 at an early post zygotic stage (Figure 2). Loss of the maternal 14 however will resolve the trisomy to a normal disomy with biparental inheritance and this is consistent with trisomy rescue in ROBs producing UPD in 50% of cases unlike the 1/3 of UPD cases for non-ROB rearrangements. In contrast, conversion of a monosomic conception resulting from fertilization of a nullisomic gamete produced by an unbalanced 2:1 adjacent segregation in the rob(13;14) mother would require replication of the paternal 14 to produce a “normal” karyotype which would be consistent with paternal iUPD 14 with two similar paternal 14s (Figure 3). It is noteworthy that the paternal homolog can also replicate as an isochromosome which would resemble a homologous ROB. All rescued monosomic conceptuses with an isochromosome would show uniparental iUPD. Most nondisjunction occurs during maternal meiosis and a nullisomic gamete when produced would involve loss of a maternal chromosome, it is therefore not unexpected that chromosomal duplication in monosomy rescue would involve paternal UPD. It must however be recognised that cases of monosomy rescue in contrast to trisomy rescue are much less common as loss of an autosome in monosomic conceptuses are more likely to be lethal and lost in early gestation (9). Opportunities for such rescues are limited.
Gamete complementation, a much rarer UPD mechanism is a consequence of an unusual combination of meiotic errors in both parents whereby a nullisomic gamete in one parent complements a disomic gamete in the other parent producing a disomic conceptus. One such example was reported in a 9-year-old girl with paternal hUPD for chromosome 14 and a 45,XX,rob(13;14)pat karyotype. The father had a balanced ROB [45,XY,rob(13q;14q)] and the mother a balanced reciprocal translocation between the long arms of chromosomes 1 and 14 [46,XX,t(1;14)(q32;q32)]. The UPD 14pat in the proband was assessed as derived from fertilization of a nullisomic ovum with a missing 14 (resulting from a 3:1 segregation in the mother) by a disomic sperm that had both the rob(13q;14q) and a normal 14 (14).
Genomic imprinting and clinically significant UPD phenotypes involving the acrocentric chromosomes
Genomic imprinting refers to the differential expression of a gene depending on its parent of origin and is revealed in cases of hetero/iUPD. Some genes are expressed preferentially from the maternal or paternal alleles and depending on which parental contribution, the difference in gene expression would affect the subsequent phenotype. Genes in a region subjected to imprinting will have only one copy of the gene expressed while the other is silenced.
Amongst the acrocentric chromosomes, chromosomes 14 and 15 have been established as imprinted with defined clinical phenotypes. UPD 14 (mat and pat) and UPD 15mat/pat (PWS/AS) have been revealed in familial or de novo ROB cases. In familial balanced ROBs transmitted from parent to child, UPD derived from chromosomes 14 and 15 has been determined in both a setting of a balanced ROB involving these two chromosomes with each other and with other acrocentrics and though uncommon, also in a normal karyotype (23).
UPD 14mat
Maternal UPD for chromosome 14 was first described in a 17-year-old male with a balanced rob(13q;14q) who inherited the single free chromosome 14 and the rob(13q;14q) from the mother consistent with “correction” of a rob(13q;14q)+14 conception (15). Patients with UPD 14mat have been described to have pre- and post-natal growth retardation, hypotonia, small hands and feet, scoliosis, premature puberty and normal to mild developmental delay (15,24). UPD 14mat have been reported in association with ROBs, isochromosomes and mosaicism.
UPD 14pat
A more severe phenotype is seen in UPD 14pat patients with dysmorphic facial features, short limbs, narrow thorax with short abnormally curved ribs described as “coat-hanger”-like, congenital heart defects and mental retardation (25). The clinical picture with thoracic deformity secondary to rib abnormalities is not unlike that of segmental UPD for 14q32 (26) and supports the critical segment relevant to the phenotype to be at 14q32.2 with imprinted loci. These include paternally expressed genes DLK1 (delta, Drosophila homologue-like1) and RTL1 and maternally expressed genes MEG3 (maternally expressed gene 3)/GTL2 (gene trap locus 2) and RTL1 (RTL1 antisense) (27). An 8-month-old girl with a normal 46,XX karyotype whose mother is a rob(13q;14q) carrier was determined to have UPD 14pat. The proposed mechanism for this rare case involves a nullisomic gamete from maternal MI nondisjunction producing a monosomic conceptus “rescued” by duplication of the paternal 14 (23).
UPD 15mat and PWS (PWS, OMIM 176270)
PWS is characterised by neonatal hypotonia, feeding difficulties in early infancy, facial dysmorphism, obesity, developmental delay, short stature and/or mental retardation. The PWS critical region lies within a 2.5 Mb differentially imprinted region on the proximal long arm of chromosome 15. PWS is caused by a lack of transcript involving imprinted genes, MKRN3, MAGEL2, NECDIN, SNURF-SNRPN, C15orf2 and a cluster of small nucleolar RNAs (snoRNAs) on the paternal chromosome (28). Several family studies have revealed the SNORD116 snoRNA gene cluster critical to the phenotype (12). The maternally inherited copies of these genes remain silent due to imprinting and only the paternal copies of the genes are expressed. Loss of function of paternal copies of this region results in PWS.
Maternal UPD15 represents only about 20-30% of PWS patients, the majority of patients (about 65-75%) having a deletion of 15q12 on the paternally derived chromosome. A third class of PWS patients involving 1-3% of cases have biparental inheritance and an imprinting centre defect that renders the paternal contribution non-functional (28). UPD 15mat is more commonly derived from correction of a trisomic conceptus and usually presents as a normal 46,XX or 46,XY karyotype and in situations involving ROBs, a balanced ROBs involving a 15. UPDs as such are generally associated with MI non-disjunction in older mothers and a five-fold increase in those showing UPD have been born to mothers 35 years and over (29). UPD individuals compared to deletion patients are more likely to develop psychosis and have autism spectrum disorders (30).
If the mother has a ROB involving 15, trisomy rescue will lead to PWS and if the father has a 15 derived ROB, monosomy rescue will lead to PWS.
UPD 15pat and AS (AS, OMIM 105830)
AS is a rare neurodevelopmental disorder, characterised by ataxia, jerky limb movements, seizures, mental and developmental delay and a happy laughing disposition. Proximal 15q has two adjacent imprinted domains, one on the paternal 15 and a more distal complex of alleles on the maternal 15. Recognised disease mechanisms in AS include a maternally derived deletion of 15q11-q13 (about 70%), paternal UPD15 (2-5%), imprinting centre defect affecting the maternal 15 (2-4%) and point mutations (about 10%) in the UBE3A (E6-AP ubiquitin-protein ligase) gene (12). Imprinted UBE3A expression is restricted to brain cells and disruption of its expression (absence of gene activity) in the maternal chromosome 15 is considered to be the major cause of the disease phenotype (12).
UPD patients in AS have a less severe phenotype than deletion patients and they have less ataxia and seizures and a better development (31). Observations have been made that most paternal UPD cases have been associated with iUPD and various examples have been described in paternal UPD for chromosomes 14 and 15 (32-34).
UPD due to a parental ROB is rare and prenatal studies have shown a risk of 0.6-0.8% (35) for non-homologous ROBs. If a familial non-homologous ROB involving a chromosome 15 in a male carrier is shown to give rise to a trisomic 15 conceptus with subsequent post-zygotic loss of the maternal 15, a paternal hUPD for AS is seen. Both 15s are derived from the father and neither chromosome 15 expresses the AS critical region on the maternal 15. However, the reverse is also possible with “monosomy rescue”. As most cases involve iUPD from a postzygotic origin of the extra paternal 15, presumably “correction” of a monosomy 15 due to a nullisomic ovum followed by post zygotic origin of the extra paternal chromosome is the more likely mechanism (9). This maternal age effect and the less occurring monosomic conception requiring rescue is consistent with the finding of reduced numbers of UPD cases in AS when compared to PWS.
Absence of imprinted genes on chromosomes 13, 21 and 22
UPD for the remaining three acrocentrics, chromosomes 13, 21 and 22 have no adverse clinical impact as these chromosomes are not subject to imprinting (36).
UPD 13
Reported cases of maternal and paternal UPD for chromosome 13 have been phenotypically normal with a lack of parental imprinting effect (37,38). Familial UPD 13 with no clinical significance was demonstrated in a healthy mother [i(13q)pat] and son [i(13q)mat] with transmission of an isochromosome for the long arm of chromosome 13 apparent (39).
UPD 21
UPD for chromosome 21 also appeared not to be associated with abnormal phenotypes (40). A family investigated for multiple recurrent trisomy 21 conceptions demonstrated UPD 21 in its healthy euploid members (41).
UPD 22
No significant clinical impact was demonstrated in patients with maternal and paternal UPD 22 (5,9). UPD 22mat was shown in a healthy 25 year old man karyotyped subsequent to repeated spontaneous abortions in his wife (42). He had a de novo balanced rob(22q;22q) which was later shown to be an isochromosome for chromosome 22. Apart from causing reproductive failure with possible conceptions either monosomic or trisomic for chromosome 22, no adverse phenotypic effect was apparent.
Risks for UPD associated with ROBs
Carriers of nonhomologous ROBs are at increased risk for offspring with UPD. Risk estimates have been based on limited empirical data and interpretation from collective figures from different surveys can be made difficult with some studies including all acrocentrics involved in the ROB while others considered only ROBs that contain the clinically relevant imprinted chromosomes 14 and 15 (35). However, cumulative data from seven separate prenatal studies of reports published during the period from 2000 to 2004 revealed 4 out of 482 ROB carriers to have UPD with a risk of 0.8% (95% CI, 0.3-2.1%) (35). This is not unlike the 0.6% (95% CI, 0.01-3.3%) risk reported in one of the earlier prenatal surveys in which one UPD 13 was found amongst 168 nonhomologous ROBs (43). When only chromosomes 14 and 15 were considered, the risk of UPD was 0.6% (95% CI, 0.2-1.7%) (35). It was suggested that the 0.8% risk of UPD from pooled data reported in (35) can be applicable to all three classes of inheritance of the ROB, maternal, parental and de novo as the frequency figures shown across the three ROB groups were not statistically significant (9,35). Breakdown frequency figures include two UPD cases (UPD 13mat and UPD 14mat) among 201 maternally inherited ROBs and none in the 170 paternally inherited group. Two of the 97 de novo cases were shown to have UPD 14 from a rob(13q;14q) and a rob(14q;21q) The sample numbers were small and it may not be appropriate at this stage to attribute a higher UPD risk estimate to de novo ROB cases until larger surveys are available. De novo nonhomologous ROBs have been recommended for prenatal UPD testing as postnatal reports of UPD has been indicated in such de novo cases (44). It is obvious too that in view of the preferential maternal origin of malsegregation non-disjunction events and the effects of advanced maternal age, to presume a higher UPD risk for female Rob carriers is yet to be supported by more data. The UPD risk for paternally derived ROBs though low is certainly not negligible as UPD cases investigated postnatally because of abnormal phenotypes have been paternally associated (14,45).
There is less data on fetuses with a normal karyotype conceived from a ROB carrier parent. The risk is predicted to be low and expected to be less than that for a ROB carrier fetus. This is because the mechanism required involves monosomy rescue which is less commonly observed as monosomic conceptions tend to be fairly lethal in early gestation allowing less opportunity for a duplication event to occur. No UPD was detected in two prenatal studies involving a total of 36 karyotypically normal fetuses from nonhomologous ROB parents (46,47). The risk however is not negligible as UPD 14pat has been described in a 8-month-old girl with a normal karyotype and a carrier mother with a balanced rob(13q;14q) (23). This would suggest a need for prenatal testing to be considered. A duplication of paternal chromosome 14 consistent with monosomy rescue has been proposed as a possible mechanism.
Over a seven year period from 2007 to 2013, our laboratory had 18 fetuses prenatally diagnosed with balanced nonhomologous ROBs involving chromosomes 14 and 15, tested for UPD. They include six de novo and twelve familial (9 inherited through the mother and 3 through the father) cases. The results were unremarkable with all cases showing biparental inheritance and negative for UPD. This finding is not unlike the earlier small studies published (48,49). It is generally accepted that families with a fetus carrying a nonhomologous ROB may be counselled that the UPD risk is <1%.
In prenatal diagnosis of ROBs, the risk of UPD is also dependent on whether the ROB is heterologous or homologous. In a study of 174 prenatally studied ROBs, four of the six homologous translocations identified were positive for UPD providing a risk estimate of 66% (43). It is known that most rearrangements that resemble homologous ROBs are isochromosomes and not true ROBs which are derived from the post fertilization fusion of maternal and paternal homologs and reflecting biparental inheritance. These four ROBs were de novo and subsequently identified as isochromosomes. The two remaining homologous ROBs were UPD negative and confirmed as true homologous ROBs. As is generally recognised, all isochromosomes and some homologous ROBs will display iUPD and a clinically adverse phenotype would present if the imprintable chromosomes 14 and 15 or involved. These cases would also carry a small risk of homozygosity for recessive mutations and recessive disease. Detection of a homologous ROB involving the imprinted chromosomes 14 and 15 would be of concern due to their possible isochromosome nature and risk of iUPD. Of interest were two prenatally detected homologous rob(15q;15q) showing biparental inheritance resulting from postzygotic ROB fusion between both parental chromosome 15 (50,51). The reproductive potential of an individual who carry a homologous rob(15q;15q) would mainly include repeated miscarriages consistent with trisomy or monosomy 15. Apart from this, should trisomy rescue occur in a trisomy 15 conception and should the individual be male, a child with AS and UPD 15pat would be the outcome in contrast to PWS and UPD 15mat from a female rob(15;15q) carrier.
Recommendations on UPD testing
Given the relevance of the imprinted chromosomes 14 and 15, ROBs involving these chromosomes would have a relatively higher possibility of resulting in UPD. UPD analysis should be offered to couples in the prenatal diagnosis of ROBs as it provides no additional risk to the pregnancy. This is recognised in the recommendations of both the American and Canadian College of Medical Genetics on the diagnostic testing of UPD (52,53).
Indications for prenatal and postnatal UPD testing involving ROBs
Guidelines from the American and Canadian College of Medical Genetics (52,53) for pre/postnatal UPD testing include:
Prenatal UPD testing
Prenatal UPD testing would provide an indication of the risk of an imprinting disorder and it is advised that it be pursued in a timely manner with appropriate time for decision making and counselling.
UPD testing is recommended in fetuses with:
- A familial or de novo balanced ROB (or isochromosome) involving a chromosome 14 or 15;
- A normal karyotype when the parent is a carrier of a balanced ROB with chromosomes 14 and/or 15 [The samples studied with a normal karyotype from a ROB carrier parent are low and a significant risk is not indicated. However a recently described case (23) merits attention for UPD testing in this category];
- Fetuses with anomalies identified by ultrasound that are consistent with features found in UPD syndromes.
Postnatal UPD testing
This is required to provide a diagnosis for individuals and facilitate patient management. Parents would need to be informed of prognosis and risk estimates.
Postnatal UPD testing is recommended for:
- Individuals who present with multiple congenital anomalies, developmental delay/mental retardation and who have either a familial or de novo balanced ROB involving chromosome 14 or 15;
- Patients with clinical features suggestive of maternal and paternal UPD14, PWS and AS.
The recurrence risk for UPD is thought to be low and is largely unknown, a <1% recurrence risk had been estimated for the UPD of chromosome 15 pertinent to PWS and AS (12).
Testing methodologies—DNA polymorphism studies
DNA polymorphism analysis using short tandem repeats (STR) is still the preferred test to diagnose UPD particularly in cases of balanced rearrangements like ROBs. Testing should be performed on DNA collected from parents, child/fetus using at least two fully informative polymorphic markers.
Conclusions
ROBs confer a risk of aneuploidy. Numerous reports of UPD caused by the imprinted chromosomes 14 and 15 would suggest that phenotypically abnormal carriers as well as prenatal cases demonstrating ROBs involving these two chromosomes should be considered for UPD testing. Fetuses with a normal karyotype when a parent is a balanced ROB carrier of chromosomes 14 and/or 15 should also be included.
Acknowledgements
The author wishes to thank Dr. Michael Buckley, Clinical Director, SEALS Genetics Laboratory, NSW Health Pathology, Prince of Wales Hospital, for reviewing the manuscript. Thanks also to Dr. Bruce Bennetts and staff of Molecular Genetics, The Children’s Hospital, Westmead for the UPD DNA polymorphism studies.
Footnote
Conflicts of Interest: The author has no conflicts of interest to declare.
References
- Engel E. A new genetic concept: uniparental disomy and its potential effect, isodisomy. Am J Med Genet 1980;6:137-43. [PubMed]
- Spence JE, Perciaccante RG, Greig GM, et al. Uniparental disomy as a mechanism for human genetic disease. Am J Hum Genet 1988;42:217-26. [PubMed]
- Shaffer LG. Uniparental disomy: mechanisms and clinical consequences. Fetal Matern Med Rev 2003;14:155-75.
- Liehr T. Cytogenetic contribution to uniparental disomy (UPD). Mol Cytogenet 2010;3:8. [PubMed]
- Liehr T. Cases with uniparental disomy (UPD). Available online: http://www.med.uni-jena.de/fish/sSMC/00START-UPD.htm
- Hamerton JL, Canning N, Ray M, et al. A cytogenetic survey of 14,069 newborn infants: incidence of chromosome abnormalities. Clin Genet 1975;8:223-43. [PubMed]
- Bandyopadhyay R, Heller A, Knox-DuBois C, et al. Parental origin and timing of de novo Robertsonian translocation formation. Am J Hum Genet 2002;71:1456-62. [PubMed]
- Therman E, Susman B, Denniston C. The nonrandom participation of human acrocentric chromosomes in Robertsonian translocations. Ann Hum Genet 1989;53:49-65. [PubMed]
- Gardner RJ, Sutherland GR, Shaffer LG. Chromosome abnormalities and genetic counseling. Oxford University Press, 2012.
- Robinson WP. Mechanisms leading to uniparental disomy and their clinical consequences. Bioessays 2000;22:452-9. [PubMed]
- Stalker HJ, Williams CA. Genetic counseling in Angelman syndrome: the challenges of multiple causes. Am J Med Genet 1998;77:54-9. [PubMed]
- Buiting K. Prader-Willi syndrome and Angelman syndrome. Am J Med Genet C Semin Med Genet 2010;154C:365-76. [PubMed]
- Nicholls RD, Knoll JH, Butler MG, et al. Genetic imprinting suggested by maternal heterodisomy in nondeletion Prader-Willi syndrome. Nature 1989;342:281-5. [PubMed]
- Wang JC, Passage MB, Yen PH, et al. Uniparental heterodisomy for chromosome 14 in a phenotypically abnormal familial balanced 13/14 Robertsonian translocation carrier. Am J Hum Genet 1991;48:1069-74. [PubMed]
- Temple IK, Cockwell A, Hassold T, et al. Maternal uniparental disomy for chromosome 14. J Med Genet 1991;28:511-4. [PubMed]
- Kotzot D. Review and meta-analysis of systematic searches for uniparental disomy (UPD) other than UPD 15. Am J Med Genet 2002;111:366-75. [PubMed]
- Ogur G, Van Assche E, Vegetti W, et al. Chromosomal segregation in spermatozoa of 14 Robertsonian translocation carriers. Mol Hum Reprod 2006;12:209-15. [PubMed]
- Munné S, Escudero T, Sandalinas M, et al. Gamete segregation in female carriers of Robertsonian translocations. Cytogenet Cell Genet 2000;90:303-8. [PubMed]
- Robinson WP, Bernasconi F, Mutirangura A, et al. Nondisjunction of chromosome 15: origin and recombination. Am J Hum Genet 1993;53:740-51. [PubMed]
- Purvis-Smith SG, Saville T, Manass S, et al. Uniparental disomy 15 resulting from “correction” of an initial trisomy 15. Am J Hum Genet 1992;50:1348-50. [PubMed]
- Cassidy SB, Lai LW, Erickson RP, et al. Trisomy 15 with loss of the paternal 15 as a cause of Prader-Willi syndrome due to maternal disomy. Am J Hum Genet 1992;51:701-8. [PubMed]
- Martin RH, Ko E, Rademaker A. Distribution of aneuploidy in human gametes: comparison between human sperm and oocytes. Am J Med Genet 1991;39:321-31. [PubMed]
- Potok O, Schlade-Bartusiak K, Perrier R, et al. Paternal uniparental isodisomy for chromosome 14 in a child with normal karyotype, resulting from malsegregation of maternal Robertsonian translocation. European Human Genetics Conference. Vienna, Austria. 2009;P03.147.
- Healey S, Powell F, Battersby M, et al. Distinct phenotype in maternal uniparental disomy of chromosome 14. Am J Med Genet 1994;51:147-9. [PubMed]
- Stevenson DA, Brothman AR, Chen Z, et al. Paternal uniparental disomy of chromosome 14: confirmation of a clinically-recognizable phenotype. Am J Med Genet A 2004;130A:88-91. [PubMed]
- Irving MD, Buiting K, Kanber D, et al. Segmental paternal uniparental disomy (patUPD) of 14q32 with abnormal methylation elicits the characteristic features of complete patUPD14. Am J Med Genet A 2010;152A:1942-50. [PubMed]
- Suzumori N, Ogata T, Mizutani E, et al. Prenatal findings of paternal uniparental disomy 14: Delineation of further patient. Am J Med Genet A 2010;152A:3189-92. [PubMed]
- Cassidy SB, Schwartz S, Miller JL, et al. Prader-Willi syndrome. Genet Med 2012;14:10-26. [PubMed]
- Ginsburg C, Fokstuen S, Schinzel A. The contribution of uniparental disomy to congenital development defects in children born to mothers at advanced childbearing age. Am J Med Genet 2000;95:454-60. [PubMed]
- Veltman MW, Thompson RJ, Roberts SE, et al. Prader-Willi syndrome--a study comparing deletion and uniparental disomy cases with reference to autism spectrum disorders. Eur Child Adolesc Psychiatry 2004;13:42-50. [PubMed]
- Fridman C, Varela MC, Kok F, et al. Paternal UPD15: further genetic and clinical studies in four Angelman syndrome patients. Am J Med Genet 2000;92:322-7. [PubMed]
- Kagami M, Nishimura G, Okuyama T, et al. Segmental and full paternal isodisomy for chromosome 14 in three patients: narrowing the critical region and implication for the clinical features. Am J Med Genet A 2005;138A:127-32. [PubMed]
- Papenhausen PR, Mueller OT, Johnson VP, et al. Uniparental isodisomy of chromosome 14 in two cases: an abnormal child and a normal adult. Am J Med Genet 1995;59:271-5. [PubMed]
- Fridman C, Koiffmann CP. Origin of uniparental disomy 15 in patients with Prader-Willi or Angelman syndrome. Am J Med Genet 2000;94:249-53. [PubMed]
- Shaffer LG. Risk estimates for uniparental disomy following prenatal detection of a nonhomologous Robertsonian translocation. Prenat Diagn 2006;26:303-7. [PubMed]
- Kotzot D, Utermann G. Uniparental disomy (UPD) other than 15: phenotypes and bibliography updated. Am J Med Genet A 2005;136:287-305. [PubMed]
- Stallard R, Krueger S, James RS, et al. Uniparental isodisomy 13 in a normal female due to transmission of a maternal t(13q13q). Am J Med Genet 1995;57:14-8. [PubMed]
- Soler A, Margarit E, Queralt R, et al. Paternal isodisomy 13 in a normal newborn infant after trisomy rescue evidenced by prenatal diagnosis. Am J Med Genet 2000;90:291-3. [PubMed]
- Slater H, Shaw JH, Bankier A, et al. UPD 13: no indication of maternal or paternal imprinting of genes on chromosome 13. J Med Genet 1995;32:493. [PubMed]
- Blouin JL, Avramopoulos D, Pangalos C, et al. Normal phenotype with paternal uniparental isodisomy for chromosome 21. Am J Hum Genet 1993;53:1074-8. [PubMed]
- Bán Z, Nagy B, Papp C, et al. Recurrent trisomy 21 and uniparental disomy 21 in a family. Fetal Diagn Ther 2003;18:454-8. [PubMed]
- Schinzel AA, Basaran S, Bernasconi F, et al. Maternal uniparental disomy 22 has no impact on the phenotype. Am J Hum Genet 1994;54:21-4. [PubMed]
- Berend SA, Horwitz J, McCaskill C, et al. Identification of uniparental disomy following prenatal detection of Robertsonian translocations and isochromosomes. Am J Hum Genet 2000;66:1787-93. [PubMed]
- Antonarakis SE, Blouin JL, Maher J, et al. Maternal uniparental disomy for human chromosome 14, due to loss of a chromosome 14 from somatic cells with t(13;14) trisomy 14. Am J Hum Genet 1993;52:1145-52. [PubMed]
- Cotter PD, Kaffe S, McCurdy LD, et al. Paternal uniparental disomy for chromosome 14: a case report and review. Am J Med Genet 1997;70:74-9. [PubMed]
- Sensi A, Cavani S, Villa N, et al. Nonhomologous Robertsonian translocations (NHRTs) and uniparental disomy (UPD) risk: an Italian multicentric prenatal survey. Prenat Diagn 2004;24:647-52. [PubMed]
- Ruggeri A, Dulcetti F, Miozzo M, et al. Prenatal search for UPD 14 and UPD 15 in 83 cases of familial and de novo heterologous Robertsonian translocations. Prenat Diagn 2004;24:997-1000. [PubMed]
- Jay AM, Roberts E, Davies T, et al. Prenatal testing for uniparental disomy (UPD). Prenat Diagn 2001;21:513. [PubMed]
- Gualandi F, Sensi A, Trabanelli C, et al. Prenatal UPD testing survey in Robertsonian translocations. Prenat Diagn 2000;20:465-8. [PubMed]
- Cheung SW, Shaffer LG, Richards CS, et al. Prenatal diagnosis of a fetus with a homologous Robertsonian translocation of chromosomes 15. Am J Med Genet 1997;72:47-50. [PubMed]
- Abrams DJ, Aronoff AR, Ann Berend S, et al. Prenatal diagnosis of a homologous Robertsonian translocation involving chromosome 15. Prenat Diagn 2001;21:676-9. [PubMed]
- Shaffer LG, Agan N, Goldberg JD, et al. American College of Medical Genetics statement of diagnostic testing for uniparental disomy. Genet Med 2001;3:206-11. [PubMed]
- Dawson AJ, Chernos J, McGowan-Jordan J, et al. CCMG guidelines: prenatal and postnatal diagnostic testing for uniparental disomy. Clin Genet 2011;79:118-24. [PubMed]


