
Personalising medicine in inflammatory bowel disease—current and future perspectives
Background
Inflammatory bowel disease (IBD) has classically been considered to comprise Crohn’s disease (CD), ulcerative colitis (UC) and IBD unclassified (IBDU). This group of chronic, relapsing and remitting inflammatory conditions primarily affect the gastrointestinal tract leading to long-term morbidity, with multiple downstream sequelae. Current management strategies focus on treating disease relapses, when they occur, and prolonging remission with immunomodulators and monoclonal antibody therapy (1-3). Up to 25% of IBD patients present during childhood (before the age of 18 years) (4,5). Surgical intervention is often required in both UC and CD, occurring in 10% and 25% of children prior to the age of 18 years respectively (6,7).
Despite increased understanding of the underlying disease process there is no cure. Many of the medications currently available are extremely effective if used as part of a timely and precise management strategy (8). However the same treatments have significant potential toxicity including increased infection risk, steroid toxicity, increased risk of malignancy, and many patients can lose response when treatments are used long term (9,10).
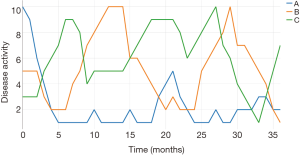
The primary aim of treatment has moved from symptom management to full mucosal and in CD, transmural healing, with the additional challenges in childhood of achieving normal growth, puberty and access to education (11). Current treatment protocols can be effective at managing patients but many children fail to respond, have frequent relapses, progress to surgery and develop significant complications. In adult-onset disease there is a move towards a top-down approach, giving monoclonal antibody therapy as initial treatment to modify the natural history of disease progression (12). However, this exposes patients, likely to require lifelong therapy, to earlier risks of serious infections, the potential for malignancy (including hepatocellular T-cell lymphoma) and secondary loss of response (13). In addition, a top-down approach is clearly not appropriate for all patients, with some having a quiet and indolent disease course requiring only minimal therapy compared to others with rapidly progressive disease. Figure 1 shows the potential disease course for three patients with IBD, highlighting these differences. The distinction of patients into these subgroups, alongside additional stratification through machine learning (ML), is the cornerstone of personalised medicine that will be discussed in this review.
What is IBD?
Personalised therapy hinges on the precise understanding of the underlying, molecular diagnosis. The characteristic feature of IBD is chronic, relapsing and remitting intestinal inflammation. Classically there have been two main subtypes, CD and UC, each with typical features pointing to that diagnosis. CD can occur from mouth to anus, is patchy, transmural, granulomatous and may have stricturing or penetrating (fistulating) features (14). Conversely UC is a disease of the colonic and rectal mucosa (potentially with limited ‘backwash’ ileitis) that does not lead to fibrotic strictures or perianal disease (15). Patients not fitting neatly into either group are described as IBDU, something which is more common in children (16). Whilst there are diagnostic criteria for each disease subtype in both adults and children it is clear that there is also vast disease heterogeneity, even within disease subtypes, with varying disease appearance and outcome (17-19). Additionally, there is frequently macroscopic/histological uncertainty and subsequent reclassification of disease, including after surgical resection (20). The underlying pathogenesis points to a complex multifactorial aetiology including genetic defects in pathways associated with the immune system, epithelial barrier, bacterial recognition and response, and environmental impacts including a dysbiotic microbiome (21,22). There is significant overlap in the genetic susceptibility to CD and UC, with cases frequently having both disease subtypes in their family history (21,23,24). In 2011 Khor et al. identified 28 risk loci through genome-wide association studies (GWAS) conferring increased risk for both CD and UC (21). A year later Jostins et al. [2012] detailed 110 of the 163 known susceptibility loci as risk for both CD and UC and most recently Liu et al. [2015] reported 38 new single nucleotide polymorphisms (SNPs), of which 27 were joint risk loci for both CD and UC (25,26). Of the 201 loci identified to date through GWAS, 148 (74%) are risk loci for both CD and UC (25,26).
Whilst there is clearly some utility in classifying disease into CD or UC at diagnosis (for some treatment decisions, prognostication and research classification) it is likely that this system is overly simplistic. Recent unsupervised approaches for classification of disease based on clinical, genetic and microbial data do not result in the two disease subtypes, instead grouping patients into multiple different categories (27-29). Moreover, it may be that each individual presenting with IBD has their own specific reason for having disease based on their underlying genetic predisposition (including immune function or epithelial barrier dysfunction) and interaction with the environment (diet, microbiome, infection). Additionally, variation must be seen in the context of subtle modifiers in interacting genes and the current diagnostic categories may be superseded by a precise molecular diagnosis.
Current management strategies for paediatric IBD
The initial aim is the safe and rapid induction of remission and optimisation of nutrition. Once remission has been established the maintenance of remission (prevention of relapse) is the priority. Treatments are summarised in Table 1.
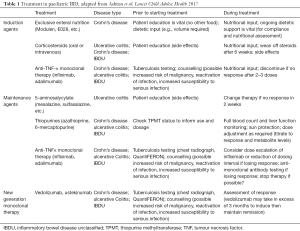
Full table
Induction of remission
Exclusive enteral nutrition (EEN)
EEN is the first line option for induction of remission in paediatric CD (2). It is taken in the form of a liquid drink for 6–8 weeks with complete exclusion of all other foods and drinks. There is an excellent response rate of up to 80%, with appropriate case selection (30). Maintenance therapy is often started during the EEN treatment course.
Corticosteroids
In moderate/severe CD the second line agent is oral steroids with consideration of intravenous therapy in severe, pan-enteric disease. Some patients will not response to EEN and will require steroids to induce remission, although the reasons behind this are often unclear (excluding patient factors and tolerability) (30,31).
Steroids (oral or intravenous) are recommended for induction of remission in UC (3). The response rate is up to 90%, usually used in conjunction with a 5-amino salicylic acid (5-ASA) preparation (mesalazine, sulfasalazine). Steroid refractory disease at induction triggers escalation of immunosuppression with monoclonal therapy being the most widely used.
Anti-TNF therapy
Anti-TNF monoclonal therapy (infliximab, adalimumab) is an effective induction agent in selected patients, but these cases can be difficult to identify at disease onset (32). These include disease failing to respond to steroid induction, stricturing, penetrating or fistulating disease and severe perianal disease (2,3). In some centres there is a ‘top-down’ therapy approach for all patients regardless of disease severity, which began in adult IBD although this is not currently recommended in children (12). The long-term impact (good and bad) of the ‘top-down’ approach is not yet known and this presents a significant challenge for clinicians considering this approach.
Maintenance therapy
There are many medications used to prevent relapses and maintain remission. All have (potentially severe) side effects and not all will tolerate each medication. The main priority of management is identification of patients who will respond and maintain remission with specific treatments, whilst avoiding these in patients who will not respond or will develop significant side effects. There is an increased risk of infection with all immunomodulator and biological therapies and this can lead result in significant morbidity.
5-ASA derivatives
Effective as a monotherapy to maintain remission in mild/moderate paediatric UC but often additional immunosuppression, such as with thiopurines, is required. No role in CD.
Thiopurines
Thiopurines (azathioprine and 6-mercaptopurine) are used for maintaining remission in both CD and UC, with a reasonable proportion achieving a sustained remission. Thiopurines can be combined with 5-ASA (in UC only) and anti-TNF medication to maintain remission. Safety is controversial with a rare but increased long term malignancy risk (lymphoproliferative disorders), with specifically increased risk in EBV infection in previously naïve patients (33).
Anti-TNF (monoclonal antibody) therapy
Anti-TNF therapy in paediatric IBD, particularly in steroid refractory disease is the largest step forward in the last 25 years. It is highly effective leading to prolonged remission, improved growth and mucosal healing (4). Response rates for infliximab are good, however some patients will not respond (primary non-responders) and some will lose response with time probably related to antibody formation (secondary loss of response) (34).
Dosing and interval can be varied but predictors for the level to induce or maintain remission are uncertain. Combination therapy with an immunomodulator (thiopurine or methotrexate) is recommended to reduce secondary loss of response (35). There is some concern that long-term anti-TNF therapy increases the risk of malignancy, specifically lymphoproliferative disorders (hepatosplenic T-cell lymphoma) however the absolute risk is small (33).
Other treatment options
Many other medications including methotrexate can provide an alternative in selected cases. Newer classes of monoclonal therapy including ustekinumab (anti-IL-12/23) and vedolizumab (anti-α4β7 integrin) are being used in paediatric disease although are more established in adult practice (36,37). Currently the use of these medications is restricted to anti-TNF failures or those with side effects (such as psoriasis). Earlier escalation would be helped by knowledge of which patients will fail therapy or develop significant toxicity on treatment.
What is personalised medicine?
Personalised (or precision) medicine is ‘a form of illness management that uses information about a person’s genes, proteins, and environment to prevent, diagnose, and treat disease’ (38). The vanguard application of personalised therapy has been oncology, specifically with tumour characterisation, tailored treatments and now immunotherapy. An early example was in breast cancer and has evolved into the routine screening of HER2 receptors and personalised use of medications such as Herceptin in HER2 positive patients, increasing survival and reducing side effects (39). The uptake of this approach in other areas of medicine has been slower and is largely related to the complexity of biomarker identification in complex, multi-faceted disease, but is now being applied to some rare conditions. A recent editorial describes personalised medicine as an ‘exciting prospect’ but the correct patients and disease must be identified in order for it to be effective (40). Personalised medicine has been brought to the fore in the United Kingdom by the 100,000 genomes project and the potential to apply genomic data to cancer and rare disease, giving patients a more personalised diagnosis (41). Different data types and techniques are summarised in Figure 2.
In a complex disease, such as IBD, the application of personalised medicine can be considered in a variety of areas (42):
- Diagnostics: providing rapid and accurate diagnosis of disease, including rare disease subtypes. Application of next-generation sequencing (NGS) and other biological data (such as metabolomics) with specific emphasis on genomics;
- Stratification: use of biomarkers and application of artificial intelligence (including ML strategies), leading to the creation of new subgroupings of disease (stratifying disease) through unsupervised and supervised approaches;
- Prognostication: providing accurate information based on data available at diagnosis (clinical and scientific) to provide an outlook for disease severity, complications and co-morbidities;
- Medication/treatment response: supervised stratification of patients into groups based on likelihood of response to therapy or likelihood of side effects based on data available at diagnosis. Development of new therapies based on unsupervised grouping of patients and biomarkers identification.
Current application of personalised medicine in IBD
On a simple level, the personalisation of medicine in IBD is already in existence. The use of specific investigations to minimise complications or to guide exact therapy has been employed in varying degrees for many years. Whilst these approaches are comparatively basic and do not constitute true predictive medicine, they are important and should be included in future modelling.
Testing for infection exposure
The importance of immunity or exposure to several infections are potentially important in determining risk with certain treatments. Establishing Epstein-Barr virus (EBV) exposure status is contentious; seronegativity is associated with a small increased risk of lymphomas in those subsequently exposed to EBV when treated with thiopurines, although the overall risk remains very low (10,43). Some clinicians will be more cautious in EBV-naïve patients, particularly males, and ECCO/ESPGHAN guidance is to test serology and avoid in acute infection, however most centres will still use a thiopurine, even in the seronegative patient group (2).
The routine screening of patients for tuberculosis (through a variety of methods) is now mandatory prior to starting an anti-TNF medication due to the risk of reactivation and disseminated disease (44). Patients with positive results will require concurrent treatment for tuberculosis.
Additional serological testing looking at immune status to infections such as varicella zoster, measles and mumps is useful to guide vaccination strategies, individual to each patient (45).
Thiopurine treatment
Prior to starting a thiopurine (either azathioprine or 6-mercaptopurine) the ability to metabolise the drug should be assessed (46). The current standard focuses on the activity of the thiopurine methyltransferase (TPMT) enzyme or a genotype of TPMT. Patients with low enzymatic activity are at increased risk of developing toxicity (myelosuppression, liver failure) and use/dose therefore needs to be tailored. TMPT is not the only enzyme associated with thiopurine metabolism and a more extensive pharmacogenomics panel has been suggested to guide therapy more accurately (46).
Anti-TNF therapy
A recent study by Dupont-Lucas and colleagues looked to stratify risk of loss of response to infliximab, retrospectively identifying isolated colonic disease as a risk factor (47). Whilst this is basic clinical information the move to using data to predict which patients will respond is important step to personalising therapy prior to more complex analyses. The ‘personalising anti-TNF therapy in Crohn’s disease’ (PANTS) study has recently reported the association between the HLA-DQA1*05 allele and the increased production of anti-TNF antibodies (hazard ratio 1.9) (48). However the clinical implication is unclear as there was no increased risk of loss of response in the group with increased antibodies (48).
The routine monitoring of anti-TNF levels and antibodies to infliximab/adalimumab constitutes precision therapy and is especially useful to guide therapy when patients experience loss of response (49). The concept of ‘treat to target’ emerged from rheumatoid arthritis management (50). In anti-TNF therapy these targets include ensuring adequate drug levels to avoid subsequent loss of response and to promote use of concurrent immunosuppression to avoid anti-drug antibody (ADA) generation (49). In the event that there is loss of response drug levels and presence or absence of ADA can determine whether dose intensification (increased dose or reduced dosing interval, low anti-TNF levels, no antibodies) will prove effective or whether there is true loss of response (low anti-TNF levels, antibodies present) requiring a change in medication (49).
Artificial intelligence and ML for personalised medicine
The latest technological advances in healthcare and biology have paved the way for the systematic collection of complex data that can be used to support more classical and routinely collected information. Usually referred to as multi-omic data, these unprecedentedly large datasets have the potential to provide information about DNA and RNA variation, protein abundance, gut microbiota and many more biological aspects to interpret with ‘mass’ clinical data. This new level of data abundance allows more in-depth analyses, increasing the chance of explaining complex traits.
The main characteristic of multi-omics data is its high dimensionality, where for each sample or patient thousands of measures are recorded (e.g., variants, proteins, metabolites, gene expression profiles). This aspect introduces a new challenge in computational medicine, the extraction and integration of meaningful information from noisy and complex data. This task requires novel methods capable of scaling with this higher complexity and simultaneously integrating the information from different data types.
ML
ML algorithms have the ability to providing the means for efficient analysis alongside interpretation of complex data and are currently applied across different areas of medicine and biology including cancer research, drug discovery, genomics and proteomics (29,51,52). Regardless of the specific application, it is possible to identify two very distinct and common tasks for which ML algorithms are employed; classification (supervised) and class discovery (unsupervised). A schematic illustration of this can be seen in Figure 3.
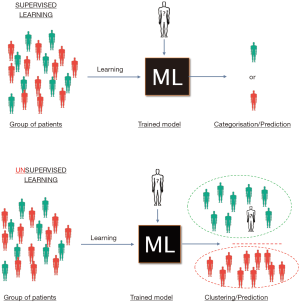
For classification purposes, supervised ML algorithms are trained against labelled (known) data and, as a result of successful learning, identify patterns that match the provided stratification. This approach has been widely chosen for the classification of patients in known disease subtypes (such as CD vs. UC) and discrimination of pathogenic and benign variants or in prognosis prediction (40,51). Concurrently, several applications of ML in cancer science have successfully classified cancer subtypes depending on histological features, genomic markers or proteomic features (51,53-55). In the context of personalised therapy, a supervised approach stratifies patients by outcomes such as response to treatment, development of complications (such as stricturing or penetrating disease), growth outcomes or requirement for escalation to monoclonal therapy/need for surgery. This model could then be used to classify patients into different risk stratifications (outcomes) at diagnosis based on patient characteristics, thus impacting on medication choices, nutritional intervention and management.
Alternatively, an unsupervised approach may be taken, allowing a ML model to group patients by how similar they are based on underlying features (such as gene variants, gene expression or microbiome signatures) and fuelling development of specific treatments for these groups based on these characteristics (including development of new medication targets). These groups may be interrogated post-hoc for enrichment of patient outcomes, such as medication response, adverse events or complications.
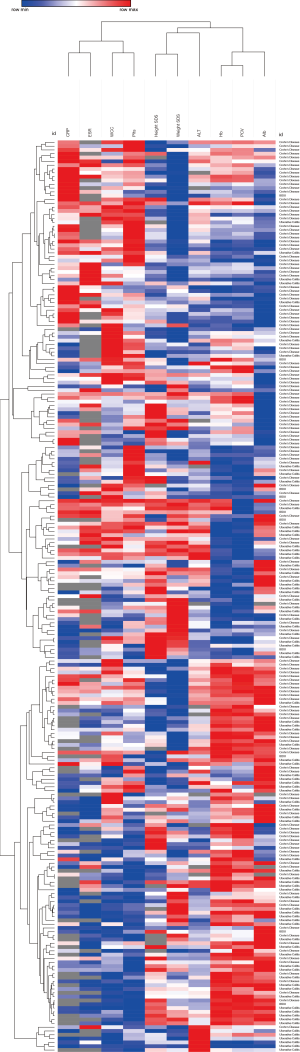
There are various mathematical tools available for ML of which support vector machines (SVMs) and random forest classifiers (RFCs) are amongst the most popular models for classification tasks. The fact that these models are relatively easy to interpret and are calculated with comparative computing efficiency has made SVM and RFCs the preferred choice for modelling biological phenomena. When the analytical question is the identification of novel patient strata, unsupervised ML tools, such as principal component analysis (PCA), multidimensional scaling (MDS) and the t-SNE algorithm, are the preferred models currently applied. An additional, and less sophisticated, tool is heirachical clustering, where samples (such as patients) can be grouped by their similarity, based on constituent features/characteristics (such as genetic or clinical data). This may reveal novel groups enriched for patients’ outcomes. An example of heirachical clustering of real data can be seen in Figure S1, merging normalised blood results with growth measures.
Despite the performance achieved by modelling single data types, there is still room for improvement by merging diverse data, achieving superior power in detecting existing and novel disease subtypes. With each data type representing a different characteristic of a single patient/individual, ML algorithms can compute simplified representations of this higher complexity. As a direct consequence, ML approaches ease the identification of novel strata, which might reflect important clinical outcomes and enable personalisation of therapy based on new groups and distinct multi-omic features.
Which data and outcomes are important for personalising therapy?
The real potential of personalised medicine lies within multi-omic data, with the potential to integrate of clinical data (bloods, outcomes, relapse, complications, etc.). Different types of multi-omic data can been seen in Figure 1. The ability to provide accurate disease stratification based on data achieved at diagnosis has precedent and is already playing a role in oncology, with the ability to predict prognosis and response to treatment with accurate biomarkers (cell surface receptors, tumour genome sequencing) providing clinically useful information to improve outcomes (56). Within IBD there are a vast range of potential therapeutic targets, based on known genetic variants and risk pathways (21). Newer therapies, including monoclonal antibodies targeting intestine specific integrin α4β7 (vedolizumab), preventing chemotaxis of inflammatory cells into the gut and anti-IL12/23 (ustekinumab) leading to specific immune regulation are likely to prove even more effective when used in target patients with disease biology sensitive to these medications (such as specific pathway disruption) (37,57). Inclusion of clinical and multi-omic data in parallel has the potential to allow assessment of the efficacy of the medication. Additional identification and in-depth phenotyping of patients who respond (or don’t respond) may be able to guide precise use in patients (58).
Defining the endpoint for therapy is an important priority, particularly for interpretation of clinical trials and observational cohort studies (59). Whilst the widely accepted target of treatment in IBD is mucosal healing (with transmural healing in CD) this is often difficult to assess in routine practice (60). Much of the published literature uses therapeutic escalation (monoclonal therapy, surgery) as a measure of disease severity. This is at least in part due to availability and reliability but also has obvious pitfalls (variation in practice between centres, individual clinician and patient choice plays a role). Predicting those patients needing rapid escalation to monoclonal therapy and those who need surgery has clear clinical application through earlier, more aggressive, therapy. Response to induction therapy is important and understanding which patients would respond to enteral nutrition or steroids in CD would be of benefit, but must account for person-specific factors (choice, palatability, personality) (30). Predicting fistulating or stricturing disease is very important and factors related to these disease types are another potential adjunct to target therapeutic endpoint. What is clear is that interpretation of results must account for the definition of a therapeutic endpoint as this is likely to be highly variable, despite ECCO/ESPGHAN published guidance (2,3).
How is personalised medicine developing in IBD and what does the future hold?
Several studies have already applied stratification techniques to patients with IBD based on clinical data (including disease location, hospitalisation records, medication usage), biomarkers (immune and molecular) and NGS in an effort to classify patients into multiple groupings based on characteristics beyond CD and UC using ML approaches (27-29,61). A summary of key paediatric studies can be seen in Table 2. A recent and large prospective study from North America applied a multi-omic approach, including clinical data to predict complications in CD (stricturing and penetrating disease), with an additional attempt to include treatment outcomes in the model (62). Whilst the competing-risk model had a specificity of 71% for predicting complications there is room for significant improvement, with an accompanying editorial discussing the limitations but also their optimism associated with the potential of personalised medicine in IBD (63).
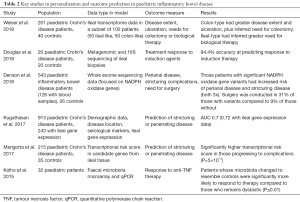
Full table
The analytical routes to clinical application can be broadly divided into two distinct approaches, as mentioned above- supervised and unsupervised classification of disease (based on multi-omic or clinical data) (Figure 2). An example of unsupervised ML can be seen in work from Weiser et al. [2018]. They report detailed gene expression profiles of adult and paediatric intestinal tissue, with two distinct clusters of CD emerging for each group. The authors performed post-hoc assessment of clinical outcomes with specific gene expression patterns, finding that colonic-type gene expression profiles were at increased risk of colectomy, whereas ileal-type were at increased risk of biological therapy (27).
Examples of early predictive modelling to stratify patient risk are now beginning to emerge. Marigorta et al. [2017] sought to forecast complicated disease based on a transcriptional risk score (a score created by the authors based on gene expression profiles) in the RISK inception cohort. The authors had a degree of success, allowing researchers to distinguish indolent versus complicating disease (stricturing or penetrating) based on the scores from 29 genes (64). A more specific approach was adopted by Denson et al. [2018] where genomic data was integrated for significant mutations in genes associated with neutrophil reactive oxygen species production, patients with these mutations had significantly increased risk of perianal and stricturing disease, providing a potential framework of aggressive treatment in this patient group from diagnosis (65). Lee et al. [2011] provided the first example of stratifying patients using an immune biomarker (transcriptional characteristics of CD8+ T cells), allowing stratification of adult patients in a high risk and low risk group for relapse (66).
Predicting response or complications related to medication is also a key area of personalised medicine (67). Two studies from Arijs et al. [2009 and 2010] analysed gene expression profiles in the mucosa of patients with CD (n=37 patients) and UC (n=46 patients). The authors found predictive, differentially expressed genes, related to response to infliximab therapy, separating responders versus non-responders with up to 95% accuracy, but these results have not since been replicated (68,69). Whilst there are fewer data on response to medications in paediatric practice, development of these models is important to predict both response to medication and equally the probability of developing side-effects and complications.
The microbiome has also been analysed in an effort to assess for factors predictive of response, an early example from Kolho et al. demonstrated microbiota returning to a similar composition to controls in paediatric patients who responded to anti-TNF therapy, but not in those who were non-responders (total number of patients treated =32). Response to anti-TNF was predicted by 6 bacterial groups (70). Recently Douglas et al. used both 16S and metagenomic sequencing to build a predictive model to identify responders to induction therapy in paediatric CD, with an accuracy of 94.4%, albeit in a cohort of only 19 paediatric patients (29). The authors found metagenomic features, in comparison to 16S sequencing, were better at classifying patients into treatment response or non-response. Shaw et al. [2016] focused on mucosal healing as an outcome for treatment, but failed to formulate a predictive model that could differentiate between ‘responders’ and ‘non-responders’ however they did identify significant differences between groups at a bacterial genus level (71). Doherty et al. [2018] observed baseline differences in the faecal microbiome (increased Faecalibacterium and Bacteroides) between CD patients responding and not responding to induction with ustekinumab therapy. They were able to predict response to therapy with an accuracy of 84.4% and concluded that microbiota may be a useful biomarker for response (72).
More recently application of clinical data has been used to predict disease outcome, complications (fibrostenosing disease, penetrating disease and perianal complications) and early relapse. Ziv-Baran et al. [2018] reported that the best clinical predictor of complications was early relapse (seen in 29%, compared to 9.7% who did not relapse), whereas subsequent relapse (within 1 year) was associated with disease activity at week 12 (including raised inflammatory markers and raised faecal calprotectin), more so than at diagnosis (73).
Drug development and future areas to target
Drug development for personalised therapy requires significant investment. Cancer has seen a boost in access to medications (specifically immunotherapy) allowing a better ‘treatment menu’ to choose from when personalising treatment (74). Over the last 10 years there have been a series of failures in development of new and effective IBD monoclonal antibody therapies against IL17 and IL13, alongside the failure to effectively use IL10 to modulate the immune response directly. All have been despite compelling evidence that these cytokines are involved in the inflammatory response in IBD (75-77). Ongoing development of JAK and SMAD7 inhibitors are showing more promise, alongside new anti-inflammatory cell trafficking (similar to vedolizumab) (59). Tofacitinib (JAK inhibitor) is now licenced for adult use in some areas. Allogeneic hematopoietic stem cell transplantation represents a truly personalised and potentially curative, established, option for a small number of children with very early onset, monogenic, IBD but is not suitable for most cases (78).
Drug development is now moving into a systems biology approach, using the understanding of underlying disease pathogenesis in this heterogeneous condition to identify potential drug targets. Response may differ between specific patient groups, including groups defined by genetics that are risk for specific problems such as surgery or stricturing disease (NOD2 and TNFSF15 mutations, respectively) (79). Additional efforts to develop therapies that target the host immune system, such as regulatory T-cells (Tregs) or through an allogenic stem cell transplant (to ‘reset’ the host immune system) appeared to show promise but progress has stalled and would be suitable for a subset of patients only (80,81).
Development of therapy based around an individual’s underlying genetic or microbial cause for disease would see personalised therapy move into a new era for IBD, with a greater selection of medications to match the underlying problem leading to inflammation in an individual.
Nutritional intervention for at-risk individuals presents another potential application. It is well established that some children with CD continue to have persistent growth deficits despite effective medical therapy (82). Simultaneously, some children will gain excess weight, becoming obese and gaining the associated medical issues and risks in later life (83). Prediction of which patients require additional nutritional support (such as in the form of supplementation), as opposed to those who do not, would provide the means to avoid persistent malnutrition, promote normal growth, prevent overnutrition and increase the likelihood that immunosuppressive and biological therapy is effective.
Conclusions
The advance of scientific discovery must be seen in the context of improving clinical care, outcomes and prognosis for children with paediatric IBD. Significant funding has been committed to IBD research, including a variety of multi-omic approaches. Whilst this has seen substantial advances in the understanding of disease the full potential of these data has not yet been fully realised, with future benefits likely to be translatable to the clinic. Specifically, the application of genetic discoveries to provide routine patient benefit is becoming possible and will hopefully be realised in the next 10 years. The potential of personalised therapy in IBD presents a tangible but significant advance in management strategy. Translating the science of precision medicine to a complex, multi-factorial, disease such as IBD presents new challenges, compared to the blueprint of cancer, but is nearing reality. A paradigm shift to personalised therapy is likely to be the next big clinical advance in paediatric IBD.
Acknowledgements
JJ Ashton is funded by an Action Medical Research, research training fellowship.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Dignass A, Van Assche G, Lindsay JO, et al. The second European evidence-based Consensus on the diagnosis and management of Crohn’s disease: Current management. J Crohns Colitis 2010;4:28-62. [Crossref] [PubMed]
- Ruemmele FM, Veres G, Kolho KL, et al. Consensus guidelines of ECCO/ESPGHAN on the medical management of pediatric Crohn’s disease. J Crohns Colitis 2014;8:1179-207. [Crossref] [PubMed]
- Turner D, Levine A, Escher JC, et al. Management of pediatric ulcerative colitis: joint ECCO and ESPGHAN evidence-based consensus guidelines. J Pediatr Gastroenterol Nutr 2012;55:340-61. [Crossref] [PubMed]
- Oliveira SB, Monteiro IM. Diagnosis and management of inflammatory bowel disease in children. BMJ 2017;357:j2083. [Crossref] [PubMed]
- Ponder A, Long MD. A clinical review of recent findings in the epidemiology of inflammatory bowel disease. Clin Epidemiol 2013;5:237-47. [PubMed]
- Ashton JJ, Versteegh HP, Batra A, et al. Colectomy in pediatric ulcerative colitis: A single center experience of indications, outcomes, and complications. J Pediatr Surg 2016;51:277-81. [Crossref] [PubMed]
- Blackburn SC, Wiskin AE, Barnes C, et al. Surgery for children with Crohn’s disease: indications, complications and outcome. Arch Dis Child 2014;99:420-6. [Crossref] [PubMed]
- Mosli M, Sabbahi H, Alyousef H, et al. Risk Stratification of Patients with Crohn’s Disease: A Retrospective Analysis of Clinical Decision Making and Its Impact on Long-Term Outcome. Dig Dis 2018;36:49-55. [Crossref] [PubMed]
- Hyams JS, Lerer T, Mack D, et al. Outcome following thiopurine use in children with ulcerative colitis: a prospective multicenter registry study. Am J Gastroenterol 2011;106:981-7. [Crossref] [PubMed]
- Gordon J, Ramaswami A, Beuttler M, et al. EBV Status and Thiopurine Use in Pediatric IBD. J Pediatr Gastroenterol Nutr 2016;62:711-4. [Crossref] [PubMed]
- Ashton JJ, Ennis S, Beattie RM. Early-onset paediatric inflammatory bowel disease. Lancet Child Adolesc Health 2017;1:147-58. [Crossref] [PubMed]
- D’Haens GR. Top-down therapy for IBD: rationale and requisite evidence. Nat Rev Gastroenterol Hepatol 2010;7:86-92. [Crossref] [PubMed]
- Hyams J, Walters TD, Crandall W, et al. Safety and efficacy of maintenance infliximab therapy for moderate-to-severe Crohn’s disease in children: REACH open-label extension. Curr Med Res Opin 2011;27:651-62. [Crossref] [PubMed]
- O’Donoghue DP, Dawson AM. Crohn’s disease in childhood. Arch Dis Child 1977;52:627-32. [Crossref] [PubMed]
- Danese S, Fiocchi C. Ulcerative Colitis. N Engl J Med 2011;365:1713-25. [Crossref] [PubMed]
- Levine A, Koletzko S, Turner D, et al. ESPGHAN revised porto criteria for the diagnosis of inflammatory bowel disease in children and adolescents. J Pediatr Gastroenterol Nutr 2014;58:795-806. [PubMed]
- Levine A, Turner D, Pfeffer Gik T, et al. Comparison of outcomes parameters for induction of remission in new onset pediatric Crohn’s disease: evaluation of the porto IBD group ‘growth relapse and outcomes with therapy’ (GROWTH CD) study. Inflamm Bowel Dis 2014;20:278-85. [Crossref] [PubMed]
- Piekkala M, Pakarinen M, Ashorn M, et al. Long-term outcomes after surgery on pediatric patients with Crohn disease. J Pediatr Gastroenterol Nutr 2013;56:271-6. [Crossref] [PubMed]
- Cameron FL, Gerasimidis K, Papangelou A, et al. Clinical progress in the two years following a course of exclusive enteral nutrition in 109 paediatric patients with Crohn’s disease. Aliment Pharmacol Ther 2013;37:622-9. [Crossref] [PubMed]
- Moum B, Ekbom A, Vatn MH, et al. Inflammatory bowel disease: re-evaluation of the diagnosis in a prospective population based study in south eastern Norway. Gut 1997;40:328-32. [Crossref] [PubMed]
- Khor B, Gardet A, Xavier RJ, et al. Genetics and pathogenesis of inflammatory bowel disease. Nature 2011;474:307-17. [Crossref] [PubMed]
- Gevers D, Kugathasan S, Denson LA, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 2014;15:382-92. [Crossref] [PubMed]
- Childers RE, Eluri S, Vazquez C, et al. Family history of inflammatory bowel disease among patients with ulcerative colitis: A systematic review and meta-analysis. J Crohns Colitis 2014;8:1480-97. [Crossref] [PubMed]
- Cleynen I, Boucher G, Jostins L, et al. Inherited determinants of Crohn’s disease and ulcerative colitis phenotypes: a genetic association study. Lancet 2016;387:156-67. [Crossref] [PubMed]
- Liu JZ, Sommeren S, van , Huang H, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015;47:979. [Crossref] [PubMed]
- Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491:119-24. [Crossref] [PubMed]
- Weiser M, Simon JM, Kochar B, et al. Molecular classification of Crohn’s disease reveals two clinically relevant subtypes. Gut 2018;67:36-42. [Crossref] [PubMed]
- Mossotto E, Ashton JJ, Coelho T, et al. Classification of Paediatric Inflammatory Bowel Disease using Machine Learning. Sci Rep 2017;7:2427. [Crossref] [PubMed]
- Douglas GM, Hansen R, Jones CMA, et al. Multi-omics differentially classify disease state and treatment outcome in pediatric Crohn’s disease. Microbiome 2018;6:13. [Crossref] [PubMed]
- Ashton JJ, Gavin J, Beattie RM. Exclusive enteral nutrition in Crohn’s disease: Evidence and practicalities. Clin Nutr 2018. [Epub ahead of print]. [Crossref] [PubMed]
- Afzal NA, Davies S, Paintin M, et al. Colonic Crohn’s disease in children does not respond well to treatment with enteral nutrition if the ileum is not involved. Dig Dis Sci 2005;50:1471-5. [Crossref] [PubMed]
- Hyams J, Damaraju L, Blank M, et al. Induction and maintenance therapy with infliximab for children with moderate to severe ulcerative colitis. Clin Gastroenterol Hepatol 2012;10:391-9.e1. [Crossref] [PubMed]
- Hyams JS, Dubinsky MC, Baldassano RN, et al. Infliximab Is Not Associated With Increased Risk of Malignancy or Hemophagocytic Lymphohistiocytosis in Pediatric Patients With Inflammatory Bowel Disease. Gastroenterology 2017;152:1901-14.e3. [Crossref] [PubMed]
- Roda G, Jharap B, Neeraj N, et al. Loss of Response to Anti-TNFs: Definition, Epidemiology, and Management. Clin Transl Gastroenterol 2016;7:e135. [Crossref] [PubMed]
- Hyams JS, Lerer T, Griffiths A, et al. Long-term outcome of maintenance infliximab therapy in children with Crohn’s disease. Inflamm Bowel Dis 2009;15:816-22. [Crossref] [PubMed]
- Conrad MA, Stein RE, Maxwell EC, et al. Vedolizumab Therapy in Severe Pediatric Inflammatory Bowel Disease. Inflamm Bowel Dis 2016;22:2425-31. [Crossref] [PubMed]
- Feagan BG, Sandborn WJ, Gasink C, et al. Ustekinumab as Induction and Maintenance Therapy for Crohn’s Disease. N Engl J Med 2016;375:1946-60. [Crossref] [PubMed]
- Definition of personalized medicine - NCI Dictionary of Cancer Terms - National Cancer Institute. Available online: (accessed 2 Aug 2018).https://www.cancer.gov/publications/dictionaries/cancer-terms/def/personalized-medicine
- Burstein HJ. The Distinctive Nature of HER2-Positive Breast Cancers. N Engl J Med 2005;353:1652-4. [Crossref] [PubMed]
- The Lancet. Personalised medicine in the UK. Lancet (London, England) 2018;391:e1. [Crossref] [PubMed]
- Peplow M. The 100,000 Genomes Project. BMJ 2016;353:i1757. [Crossref] [PubMed]
- Improving Outcomes Through Personalised Medicine Working at the cutting edge of science to improve patients’ lives. Available online: (accessed 2 Aug 2018).https://www.england.nhs.uk/wp-content/uploads/2016/09/improving-outcomes-personalised-medicine.pdf
- Warner B, Johnston E, Arenas-Hernandez M, et al. A practical guide to thiopurine prescribing and monitoring in IBD. Frontline Gastroenterol 2018;9:10-5. [Crossref] [PubMed]
- Hewitt RJ, Francis M, Singanayagam A, et al. Screening tests for tuberculosis before starting biological therapy. BMJ 2015;350:h1060. [Crossref] [PubMed]
- Reich J, Wasan S, Farraye FA. Vaccinating Patients With Inflammatory Bowel Disease. Gastroenterol Hepatol (N Y) 2016;12:540-6. [PubMed]
- Coelho T, Andreoletti G, Ashton JJ, et al. Genes implicated in thiopurine-induced toxicity: Comparing TPMT enzyme activity with clinical phenotype and exome data in a paediatric IBD cohort. Sci Rep 2016;6:34658. [Crossref] [PubMed]
- Dupont-Lucas C, Sternszus R, Ezri J, et al. Identifying Patients at High Risk of Loss of Response to Infliximab Maintenance Therapy in Paediatric Crohn’s Disease. J Crohns Colitis 2016;10:795-804. [Crossref] [PubMed]
- Sazonovs A, Kennedy N, Moutsianas L, et al. HLA-DQA1*05 is associated with the development of antibodies to anti-TNF therapy. bioRxiv 2018. [Crossref]
- Hendy P, Hart A, Irving P. Anti-TNF drug and antidrug antibody level monitoring in IBD: a practical guide. Frontline Gastroenterol 2016;7:122-8. [PubMed]
- Bouguen G, Levesque BG, Feagan BG, et al. Treat to target: a proposed new paradigm for the management of Crohn’s disease. Clin Gastroenterol Hepatol 2015;13:1042-50.e2. [Crossref] [PubMed]
- Vural S, Wang X, Guda C. Classification of breast cancer patients using somatic mutation profiles and machine learning approaches. BMC Syst Biol 2016;10:62. [Crossref] [PubMed]
- Pedersen BS, Quinlan AR. Who’s Who? Detecting and Resolving Sample Anomalies in Human DNA Sequencing Studies with Peddy. Am J Hum Genet 2017;100:406-13. [Crossref] [PubMed]
- Deeb SJ, Tyanova S, Hummel M, et al. Machine Learning-based Classification of Diffuse Large B-cell Lymphoma Patients by Their Protein Expression Profiles. Mol Cell Proteomics 2015;14:2947-60. [Crossref] [PubMed]
- Urdal J, Engan K, Kvikstad V, et al. Prognostic prediction of histopathological images by local binary patterns and RUSBoost. 2017 25th European Signal Processing Conference (EUSIPCO), IEEE 2017:2349-53.
- Dimitropoulos K, Barmpoutis P, Zioga C, et al. Grading of invasive breast carcinoma through Grassmannian VLAD encoding. PLoS One 2017;12:e0185110. [Crossref] [PubMed]
- Jackson SE, Chester JD. Personalised cancer medicine. Int J Cancer 2015;137:262-6. [Crossref] [PubMed]
- Lam MC, Bressler B. Vedolizumab for ulcerative colitis and Crohn’s disease: results and implications of GEMINI studies. Immunotherapy 2014;6:963-71. [Crossref] [PubMed]
- Flamant M, Roblin X. Inflammatory bowel disease: towards a personalized medicine. Therap Adv Gastroenterol 2018;11:1756283X17745029.
- Bilsborough J, Targan SR, Snapper SB. Therapeutic Targets in Inflammatory Bowel Disease: Current and Future. Am J Gastroenterol Suppl 2016;3:27-37. [Crossref]
- Dave M, Loftus E V. Mucosal healing in inflammatory bowel disease-a true paradigm of success? Gastroenterol Hepatol (N Y) 2012;8:29-38. [PubMed]
- Waljee AK, Lipson R, Wiitala WL, et al. Predicting Hospitalization and Outpatient Corticosteroid Use in Inflammatory Bowel Disease Patients Using Machine Learning. Inflamm Bowel Dis 2017;24:45-53. [Crossref] [PubMed]
- Kugathasan S, Denson LA, Walters TD, et al. Prediction of complicated disease course for children newly diagnosed with Crohn’s disease: a multicentre inception cohort study. Lancet 2017;389:1710-8. [Crossref] [PubMed]
- Arijs I, Cleynen I. RISK stratification in paediatric Crohn’s disease. Lancet 2017;389:1672-4. [Crossref] [PubMed]
- Marigorta UM, Denson LA, Hyams JS, et al. Transcriptional risk scores link GWAS to eQTLs and predict complications in Crohn’s disease. Nat Genet 2017;49:1517-21. [Crossref] [PubMed]
- Denson LA, Jurickova I, Karns R, et al. Clinical and Genomic Correlates of Neutrophil Reactive Oxygen Species Production in Pediatric Patients With Crohn’s Disease. Gastroenterology 2018;154:2097-110. [Crossref] [PubMed]
- Lee JC, Lyons PA, McKinney EF, et al. Gene expression profiling of CD8+ T cells predicts prognosis in patients with Crohn disease and ulcerative colitis. J Clin Invest 2011;121:4170-9. [Crossref] [PubMed]
- Pirmohamed M. Personalized Pharmacogenomics: Predicting Efficacy and Adverse Drug Reactions. Annu Rev Genomics Hum Genet 2014;15:349-70. [Crossref] [PubMed]
- Arijs I, Quintens R, Van Lommel L, et al. Predictive value of epithelial gene expression profiles for response to infliximab in Crohn’s disease. Inflamm Bowel Dis 2010;16:2090-8. [Crossref] [PubMed]
- Arijs I, Li K, Toedter G, et al. Mucosal gene signatures to predict response to infliximab in patients with ulcerative colitis. Gut 2009;58:1612-9. [Crossref] [PubMed]
- Kolho KL, Korpela K, Jaakkola T, et al. Fecal Microbiota in Pediatric Inflammatory Bowel Disease and Its Relation to Inflammation. Am J Gastroenterol 2015;110:921-30. [Crossref] [PubMed]
- Shaw KA, Bertha M, Hofmekler T, et al. Dysbiosis, inflammation, and response to treatment: a longitudinal study of pediatric subjects with newly diagnosed inflammatory bowel disease. Genome Med 2016;8:75. [Crossref] [PubMed]
- Doherty MK, Ding T, Koumpouras C, et al. Fecal Microbiota Signatures Are Associated with Response to Ustekinumab Therapy among Crohn’s Disease Patients. MBio 2018;9. [Crossref] [PubMed]
- Ziv-Baran T, Hussey S, Sladek M, et al. Response to treatment is more important than disease severity at diagnosis for prediction of early relapse in new-onset paediatric Crohn’s disease. Aliment Pharmacol Ther 2018;48:1242-50. [Crossref] [PubMed]
- Hunter S. Appraisal and Funding of Cancer Drugs from July 2016 (including the new Cancer Drugs Fund) A new deal for patients, taxpayers and industry. Available online: (accessed 20 Aug 2018).https://www.england.nhs.uk/wp-content/uploads/2013/04/cdf-sop.pdf
- Asadullah K, Sterry W, Volk HD. Interleukin-10 Therapy--Review of a New Approach. Pharmacol Rev 2003;55:241-69. [Crossref] [PubMed]
- Hueber W, Sands BE, Lewitzky S, et al. Secukinumab, a human anti-IL-17A monoclonal antibody, for moderate to severe Crohn’s disease: unexpected results of a randomised, double-blind placebo-controlled trial. Gut 2012;61:1693-700. [Crossref] [PubMed]
- Reinisch W, Panés J, Khurana S, et al. Anrukinzumab, an anti-interleukin 13 monoclonal antibody, in active UC: efficacy and safety from a phase IIa randomised multicentre study. Gut 2015;64:894-900. [Crossref] [PubMed]
- Uhlig HH, Schwerd T, Koletzko S, et al. The diagnostic approach to monogenic very early onset inflammatory bowel disease. Gastroenterology 2014;147:990-1007.e3. [Crossref] [PubMed]
- Cleynen I, González JR, Figueroa C, et al. Genetic factors conferring an increased susceptibility to develop Crohn’s disease also influence disease phenotype: results from the IBDchip European Project. Gut 2013;62:1556-65. [Crossref] [PubMed]
- Boden EK, Snapper SB. Regulatory T cells in inflammatory bowel disease. Curr Opin Gastroenterol 2008;24:733-41. [Crossref] [PubMed]
- Burt RK, Craig RM, Milanetti F, et al. Autologous nonmyeloablative hematopoietic stem cell transplantation in patients with severe anti-TNF refractory Crohn disease: long-term follow-up. Blood 2010;116:6123-32. [Crossref] [PubMed]
- Pfefferkorn M, Burke G, Griffiths A, et al. Growth Abnormalities Persist in Newly Diagnosed Children With Crohn Disease Despite Current Treatment Paradigms. J Pediatr Gastroenterol Nutr 2009;48:168-74. [Crossref] [PubMed]
- Gavin J, Ashton JJ, Heather N, et al. Nutritional support in paediatric Crohn’s disease: outcome at 12 months. Acta Paediatr 2018;107:156-62. [Crossref] [PubMed]



